Timothy J. Hohman
MetaVoxel: Joint Diffusion Modeling of Imaging and Clinical Metadata
Dec 12, 2025Abstract:Modern deep learning methods have achieved impressive results across tasks from disease classification, estimating continuous biomarkers, to generating realistic medical images. Most of these approaches are trained to model conditional distributions defined by a specific predictive direction with a specific set of input variables. We introduce MetaVoxel, a generative joint diffusion modeling framework that models the joint distribution over imaging data and clinical metadata by learning a single diffusion process spanning all variables. By capturing the joint distribution, MetaVoxel unifies tasks that traditionally require separate conditional models and supports flexible zero-shot inference using arbitrary subsets of inputs without task-specific retraining. Using more than 10,000 T1-weighted MRI scans paired with clinical metadata from nine datasets, we show that a single MetaVoxel model can perform image generation, age estimation, and sex prediction, achieving performance comparable to established task-specific baselines. Additional experiments highlight its capabilities for flexible inference. Together, these findings demonstrate that joint multimodal diffusion offers a promising direction for unifying medical AI models and enabling broader clinical applicability.
Pitfalls of defacing whole-head MRI: re-identification risk with diffusion models and compromised research potential
Jan 31, 2025Abstract:Defacing is often applied to head magnetic resonance image (MRI) datasets prior to public release to address privacy concerns. The alteration of facial and nearby voxels has provoked discussions about the true capability of these techniques to ensure privacy as well as their impact on downstream tasks. With advancements in deep generative models, the extent to which defacing can protect privacy is uncertain. Additionally, while the altered voxels are known to contain valuable anatomical information, their potential to support research beyond the anatomical regions directly affected by defacing remains uncertain. To evaluate these considerations, we develop a refacing pipeline that recovers faces in defaced head MRIs using cascaded diffusion probabilistic models (DPMs). The DPMs are trained on images from 180 subjects and tested on images from 484 unseen subjects, 469 of whom are from a different dataset. To assess whether the altered voxels in defacing contain universally useful information, we also predict computed tomography (CT)-derived skeletal muscle radiodensity from facial voxels in both defaced and original MRIs. The results show that DPMs can generate high-fidelity faces that resemble the original faces from defaced images, with surface distances to the original faces significantly smaller than those of a population average face (p < 0.05). This performance also generalizes well to previously unseen datasets. For skeletal muscle radiodensity predictions, using defaced images results in significantly weaker Spearman's rank correlation coefficients compared to using original images (p < 10-4). For shin muscle, the correlation is statistically significant (p < 0.05) when using original images but not statistically significant (p > 0.05) when any defacing method is applied, suggesting that defacing might not only fail to protect privacy but also eliminate valuable information.
Brain age identification from diffusion MRI synergistically predicts neurodegenerative disease
Oct 29, 2024



Abstract:Estimated brain age from magnetic resonance image (MRI) and its deviation from chronological age can provide early insights into potential neurodegenerative diseases, supporting early detection and implementation of prevention strategies. Diffusion MRI (dMRI), a widely used modality for brain age estimation, presents an opportunity to build an earlier biomarker for neurodegenerative disease prediction because it captures subtle microstructural changes that precede more perceptible macrostructural changes. However, the coexistence of macro- and micro-structural information in dMRI raises the question of whether current dMRI-based brain age estimation models are leveraging the intended microstructural information or if they inadvertently rely on the macrostructural information. To develop a microstructure-specific brain age, we propose a method for brain age identification from dMRI that minimizes the model's use of macrostructural information by non-rigidly registering all images to a standard template. Imaging data from 13,398 participants across 12 datasets were used for the training and evaluation. We compare our brain age models, trained with and without macrostructural information minimized, with an architecturally similar T1-weighted (T1w) MRI-based brain age model and two state-of-the-art T1w MRI-based brain age models that primarily use macrostructural information. We observe difference between our dMRI-based brain age and T1w MRI-based brain age across stages of neurodegeneration, with dMRI-based brain age being older than T1w MRI-based brain age in participants transitioning from cognitively normal (CN) to mild cognitive impairment (MCI), but younger in participants already diagnosed with Alzheimer's disease (AD). Approximately 4 years before MCI diagnosis, dMRI-based brain age yields better performance than T1w MRI-based brain ages in predicting transition from CN to MCI.
Multi-Modality Conditioned Variational U-Net for Field-of-View Extension in Brain Diffusion MRI
Sep 20, 2024
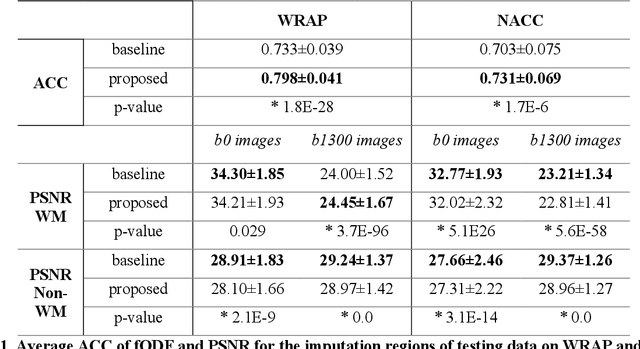

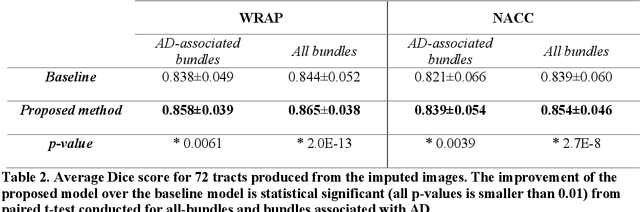
Abstract:An incomplete field-of-view (FOV) in diffusion magnetic resonance imaging (dMRI) can severely hinder the volumetric and bundle analyses of whole-brain white matter connectivity. Although existing works have investigated imputing the missing regions using deep generative models, it remains unclear how to specifically utilize additional information from paired multi-modality data and whether this can enhance the imputation quality and be useful for downstream tractography. To fill this gap, we propose a novel framework for imputing dMRI scans in the incomplete part of the FOV by integrating the learned diffusion features in the acquired part of the FOV to the complete brain anatomical structure. We hypothesize that by this design the proposed framework can enhance the imputation performance of the dMRI scans and therefore be useful for repairing whole-brain tractography in corrupted dMRI scans with incomplete FOV. We tested our framework on two cohorts from different sites with a total of 96 subjects and compared it with a baseline imputation method that treats the information from T1w and dMRI scans equally. The proposed framework achieved significant improvements in imputation performance, as demonstrated by angular correlation coefficient (p < 1E-5), and in downstream tractography accuracy, as demonstrated by Dice score (p < 0.01). Results suggest that the proposed framework improved imputation performance in dMRI scans by specifically utilizing additional information from paired multi-modality data, compared with the baseline method. The imputation achieved by the proposed framework enhances whole brain tractography, and therefore reduces the uncertainty when analyzing bundles associated with neurodegenerative.
Evaluation of Mean Shift, ComBat, and CycleGAN for Harmonizing Brain Connectivity Matrices Across Sites
Jan 24, 2024



Abstract:Connectivity matrices derived from diffusion MRI (dMRI) provide an interpretable and generalizable way of understanding the human brain connectome. However, dMRI suffers from inter-site and between-scanner variation, which impedes analysis across datasets to improve robustness and reproducibility of results. To evaluate different harmonization approaches on connectivity matrices, we compared graph measures derived from these matrices before and after applying three harmonization techniques: mean shift, ComBat, and CycleGAN. The sample comprises 168 age-matched, sex-matched normal subjects from two studies: the Vanderbilt Memory and Aging Project (VMAP) and the Biomarkers of Cognitive Decline Among Normal Individuals (BIOCARD). First, we plotted the graph measures and used coefficient of variation (CoV) and the Mann-Whitney U test to evaluate different methods' effectiveness in removing site effects on the matrices and the derived graph measures. ComBat effectively eliminated site effects for global efficiency and modularity and outperformed the other two methods. However, all methods exhibited poor performance when harmonizing average betweenness centrality. Second, we tested whether our harmonization methods preserved correlations between age and graph measures. All methods except for CycleGAN in one direction improved correlations between age and global efficiency and between age and modularity from insignificant to significant with p-values less than 0.05.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge