Derek B. Archer
Pitfalls of defacing whole-head MRI: re-identification risk with diffusion models and compromised research potential
Jan 31, 2025Abstract:Defacing is often applied to head magnetic resonance image (MRI) datasets prior to public release to address privacy concerns. The alteration of facial and nearby voxels has provoked discussions about the true capability of these techniques to ensure privacy as well as their impact on downstream tasks. With advancements in deep generative models, the extent to which defacing can protect privacy is uncertain. Additionally, while the altered voxels are known to contain valuable anatomical information, their potential to support research beyond the anatomical regions directly affected by defacing remains uncertain. To evaluate these considerations, we develop a refacing pipeline that recovers faces in defaced head MRIs using cascaded diffusion probabilistic models (DPMs). The DPMs are trained on images from 180 subjects and tested on images from 484 unseen subjects, 469 of whom are from a different dataset. To assess whether the altered voxels in defacing contain universally useful information, we also predict computed tomography (CT)-derived skeletal muscle radiodensity from facial voxels in both defaced and original MRIs. The results show that DPMs can generate high-fidelity faces that resemble the original faces from defaced images, with surface distances to the original faces significantly smaller than those of a population average face (p < 0.05). This performance also generalizes well to previously unseen datasets. For skeletal muscle radiodensity predictions, using defaced images results in significantly weaker Spearman's rank correlation coefficients compared to using original images (p < 10-4). For shin muscle, the correlation is statistically significant (p < 0.05) when using original images but not statistically significant (p > 0.05) when any defacing method is applied, suggesting that defacing might not only fail to protect privacy but also eliminate valuable information.
Brain age identification from diffusion MRI synergistically predicts neurodegenerative disease
Oct 29, 2024



Abstract:Estimated brain age from magnetic resonance image (MRI) and its deviation from chronological age can provide early insights into potential neurodegenerative diseases, supporting early detection and implementation of prevention strategies. Diffusion MRI (dMRI), a widely used modality for brain age estimation, presents an opportunity to build an earlier biomarker for neurodegenerative disease prediction because it captures subtle microstructural changes that precede more perceptible macrostructural changes. However, the coexistence of macro- and micro-structural information in dMRI raises the question of whether current dMRI-based brain age estimation models are leveraging the intended microstructural information or if they inadvertently rely on the macrostructural information. To develop a microstructure-specific brain age, we propose a method for brain age identification from dMRI that minimizes the model's use of macrostructural information by non-rigidly registering all images to a standard template. Imaging data from 13,398 participants across 12 datasets were used for the training and evaluation. We compare our brain age models, trained with and without macrostructural information minimized, with an architecturally similar T1-weighted (T1w) MRI-based brain age model and two state-of-the-art T1w MRI-based brain age models that primarily use macrostructural information. We observe difference between our dMRI-based brain age and T1w MRI-based brain age across stages of neurodegeneration, with dMRI-based brain age being older than T1w MRI-based brain age in participants transitioning from cognitively normal (CN) to mild cognitive impairment (MCI), but younger in participants already diagnosed with Alzheimer's disease (AD). Approximately 4 years before MCI diagnosis, dMRI-based brain age yields better performance than T1w MRI-based brain ages in predicting transition from CN to MCI.
Multi-Modality Conditioned Variational U-Net for Field-of-View Extension in Brain Diffusion MRI
Sep 20, 2024
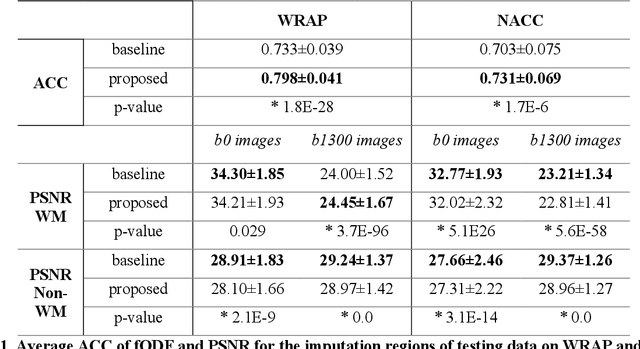

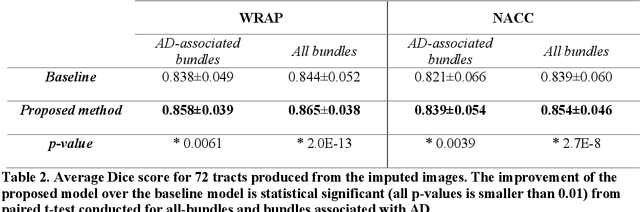
Abstract:An incomplete field-of-view (FOV) in diffusion magnetic resonance imaging (dMRI) can severely hinder the volumetric and bundle analyses of whole-brain white matter connectivity. Although existing works have investigated imputing the missing regions using deep generative models, it remains unclear how to specifically utilize additional information from paired multi-modality data and whether this can enhance the imputation quality and be useful for downstream tractography. To fill this gap, we propose a novel framework for imputing dMRI scans in the incomplete part of the FOV by integrating the learned diffusion features in the acquired part of the FOV to the complete brain anatomical structure. We hypothesize that by this design the proposed framework can enhance the imputation performance of the dMRI scans and therefore be useful for repairing whole-brain tractography in corrupted dMRI scans with incomplete FOV. We tested our framework on two cohorts from different sites with a total of 96 subjects and compared it with a baseline imputation method that treats the information from T1w and dMRI scans equally. The proposed framework achieved significant improvements in imputation performance, as demonstrated by angular correlation coefficient (p < 1E-5), and in downstream tractography accuracy, as demonstrated by Dice score (p < 0.01). Results suggest that the proposed framework improved imputation performance in dMRI scans by specifically utilizing additional information from paired multi-modality data, compared with the baseline method. The imputation achieved by the proposed framework enhances whole brain tractography, and therefore reduces the uncertainty when analyzing bundles associated with neurodegenerative.
Harmonized connectome resampling for variance in voxel sizes
Aug 02, 2024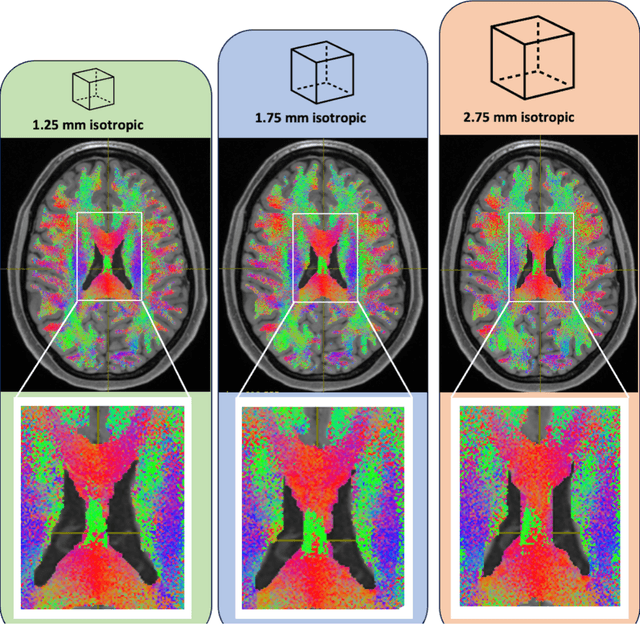
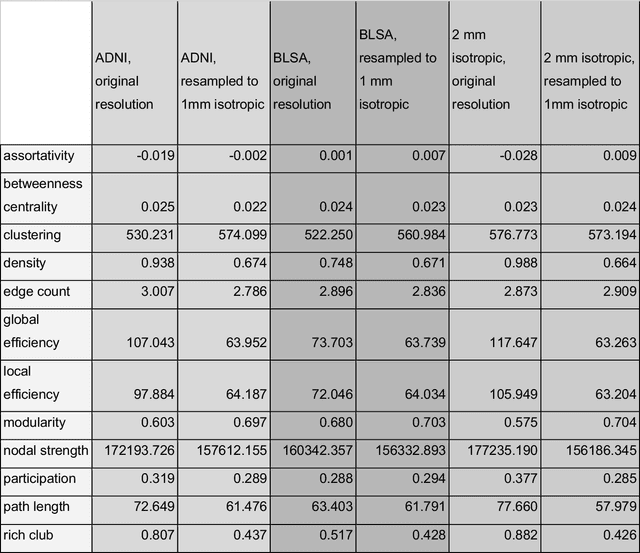
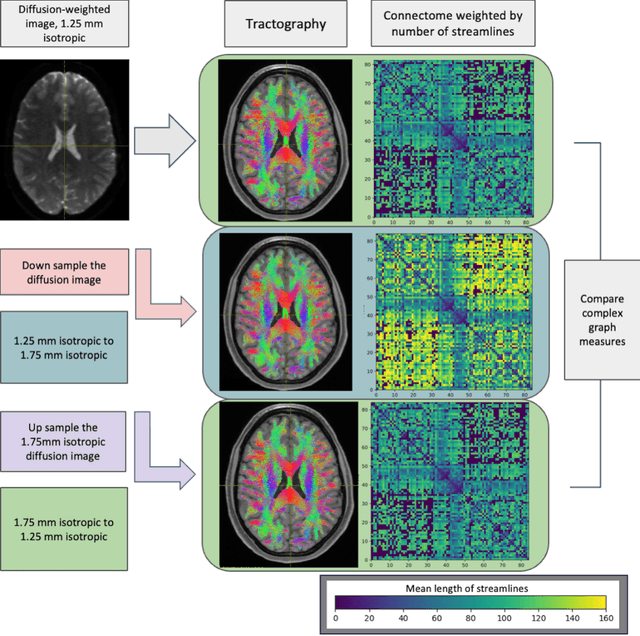
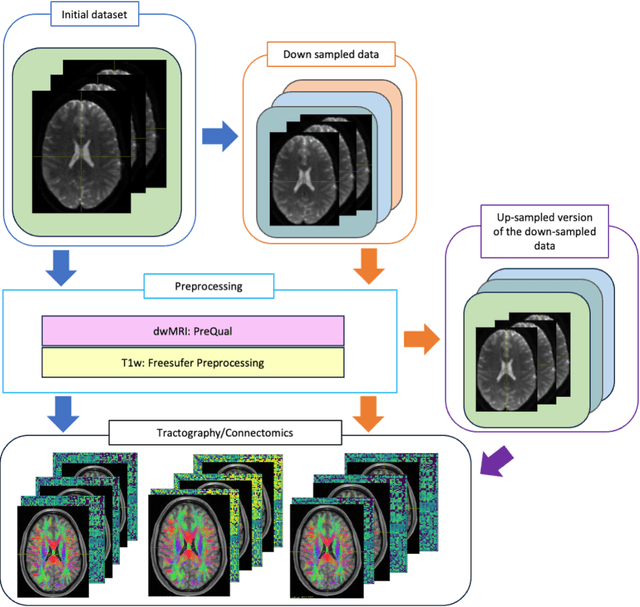
Abstract:To date, there has been no comprehensive study characterizing the effect of diffusion-weighted magnetic resonance imaging voxel resolution on the resulting connectome for high resolution subject data. Similarity in results improved with higher resolution, even after initial down-sampling. To ensure robust tractography and connectomes, resample data to 1 mm isotropic resolution.
Predicting Age from White Matter Diffusivity with Residual Learning
Nov 06, 2023Abstract:Imaging findings inconsistent with those expected at specific chronological age ranges may serve as early indicators of neurological disorders and increased mortality risk. Estimation of chronological age, and deviations from expected results, from structural MRI data has become an important task for developing biomarkers that are sensitive to such deviations. Complementary to structural analysis, diffusion tensor imaging (DTI) has proven effective in identifying age-related microstructural changes within the brain white matter, thereby presenting itself as a promising additional modality for brain age prediction. Although early studies have sought to harness DTI's advantages for age estimation, there is no evidence that the success of this prediction is owed to the unique microstructural and diffusivity features that DTI provides, rather than the macrostructural features that are also available in DTI data. Therefore, we seek to develop white-matter-specific age estimation to capture deviations from normal white matter aging. Specifically, we deliberately disregard the macrostructural information when predicting age from DTI scalar images, using two distinct methods. The first method relies on extracting only microstructural features from regions of interest. The second applies 3D residual neural networks (ResNets) to learn features directly from the images, which are non-linearly registered and warped to a template to minimize macrostructural variations. When tested on unseen data, the first method yields mean absolute error (MAE) of 6.11 years for cognitively normal participants and MAE of 6.62 years for cognitively impaired participants, while the second method achieves MAE of 4.69 years for cognitively normal participants and MAE of 4.96 years for cognitively impaired participants. We find that the ResNet model captures subtler, non-macrostructural features for brain age prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge