Synthesizing Proton-Density Fat Fraction and $R_2^*$ from 2-point Dixon MRI with Generative Machine Learning
Paper and Code
Oct 15, 2024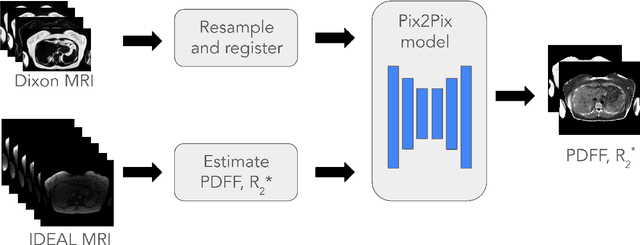



Magnetic Resonance Imaging (MRI) is the gold standard for measuring fat and iron content non-invasively in the body via measures known as Proton Density Fat Fraction (PDFF) and $R_2^*$, respectively. However, conventional PDFF and $R_2^*$ quantification methods operate on MR images voxel-wise and require at least three measurements to estimate three quantities: water, fat, and $R_2^*$. Alternatively, the two-point Dixon MRI protocol is widely used and fast because it acquires only two measurements; however, these cannot be used to estimate three quantities voxel-wise. Leveraging the fact that neighboring voxels have similar values, we propose using a generative machine learning approach to learn PDFF and $R_2^*$ from Dixon MRI. We use paired Dixon-IDEAL data from UK Biobank in the liver and a Pix2Pix conditional GAN to demonstrate the first large-scale $R_2^*$ imputation from two-point Dixon MRIs. Using our proposed approach, we synthesize PDFF and $R_2^*$ maps that show significantly greater correlation with ground-truth than conventional voxel-wise baselines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge