Anders B. Dahl
Technical University of Denmark, Visual Computing, Denmark
MMLANDMARKS: a Cross-View Instance-Level Benchmark for Geo-Spatial Understanding
Dec 19, 2025Abstract:Geo-spatial analysis of our world benefits from a multimodal approach, as every single geographic location can be described in numerous ways (images from various viewpoints, textual descriptions, and geographic coordinates). Current geo-spatial benchmarks have limited coverage across modalities, considerably restricting progress in the field, as current approaches cannot integrate all relevant modalities within a unified framework. We introduce the Multi-Modal Landmark dataset (MMLANDMARKS), a benchmark composed of four modalities: 197k highresolution aerial images, 329k ground-view images, textual information, and geographic coordinates for 18,557 distinct landmarks in the United States. The MMLANDMARKS dataset has a one-to-one correspondence across every modality, which enables training and benchmarking models for various geo-spatial tasks, including cross-view Ground-to-Satellite retrieval, ground and satellite geolocalization, Text-to-Image, and Text-to-GPS retrieval. We demonstrate broad generalization and competitive performance against off-the-shelf foundational models and specialized state-of-the-art models across different tasks by employing a simple CLIP-inspired baseline, illustrating the necessity for multimodal datasets to achieve broad geo-spatial understanding.
Deep Active Latent Surfaces for Medical Geometries
Jun 21, 2022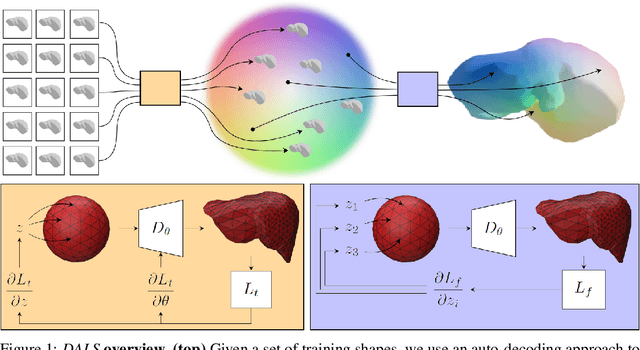
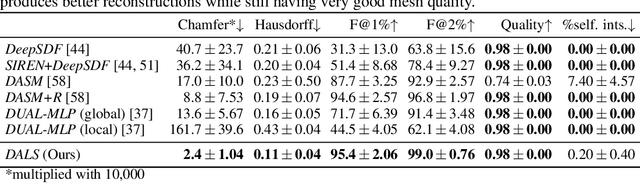
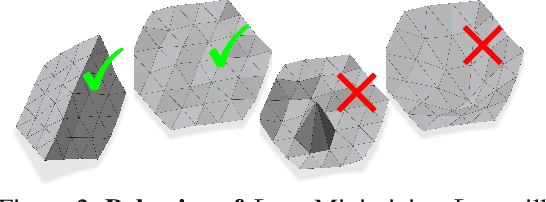
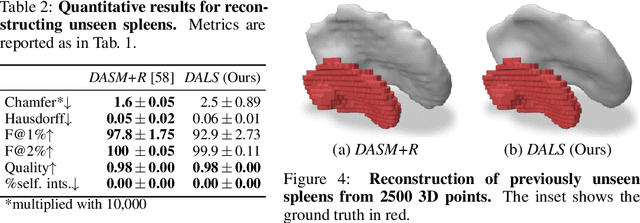
Abstract:Shape priors have long been known to be effective when reconstructing 3D shapes from noisy or incomplete data. When using a deep-learning based shape representation, this often involves learning a latent representation, which can be either in the form of a single global vector or of multiple local ones. The latter allows more flexibility but is prone to overfitting. In this paper, we advocate a hybrid approach representing shapes in terms of 3D meshes with a separate latent vector at each vertex. During training the latent vectors are constrained to have the same value, which avoids overfitting. For inference, the latent vectors are updated independently while imposing spatial regularization constraints. We show that this gives us both flexibility and generalization capabilities, which we demonstrate on several medical image processing tasks.
Review of Serial and Parallel Min-Cut/Max-Flow Algorithms for Computer Vision
Feb 01, 2022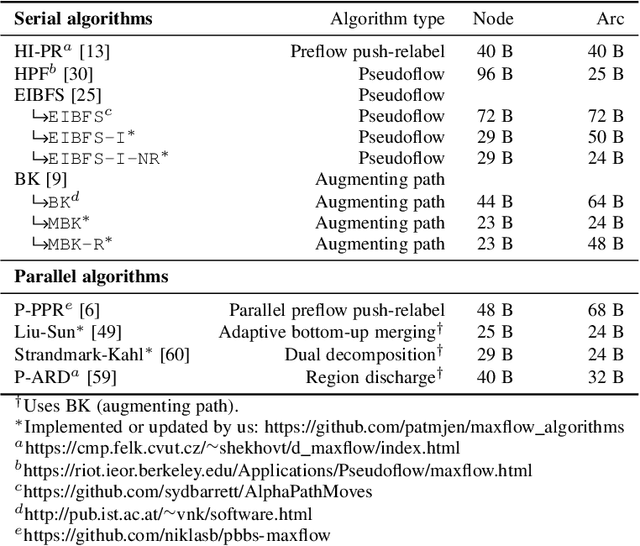
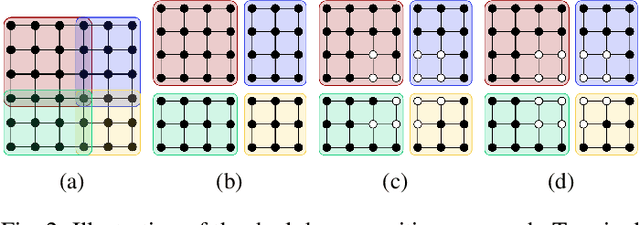
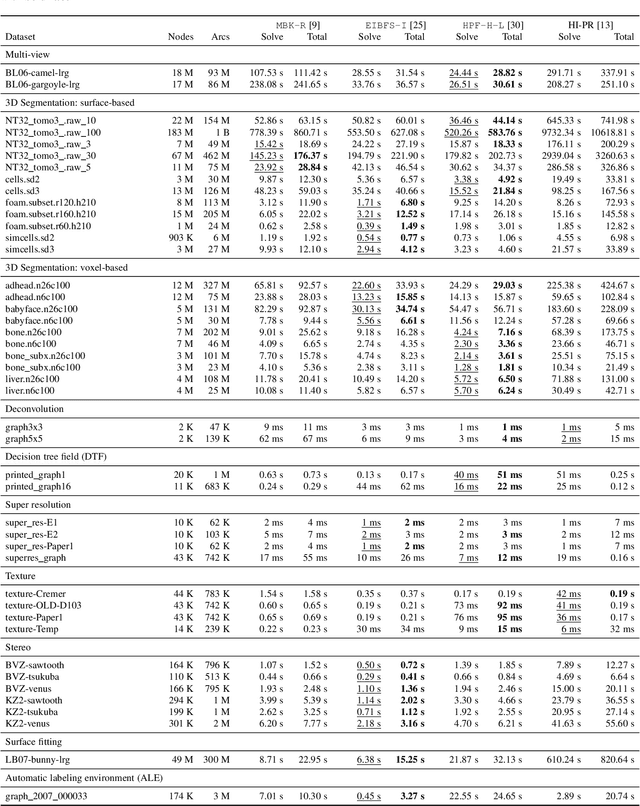
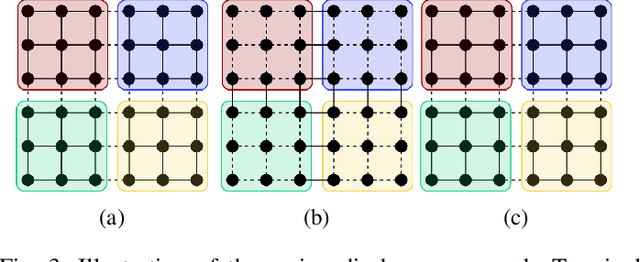
Abstract:Minimum cut / maximum flow (min-cut/max-flow) algorithms are used to solve a variety of problems in computer vision and thus significant effort has been put into developing fast min-cut/max-flow algorithms. This makes it difficult to choose an optimal algorithm for a given problem - especially for parallel algorithms, which have not been thoroughly compared. In this paper, we review the state-of-the-art min-cut/max-flow algorithms for unstructured graphs in computer vision. We evaluate run time performance and memory use of various implementations of both serial and parallel algorithms on a set of graph cut problems. Our results show that the Hochbaum pseudoflow algorithm is the fastest serial algorithm closely followed by the Excesses Incremental Breadth First Search algorithm, while the Boykov-Kolmogorov algorithm is the most memory efficient. The best parallel algorithm is the adaptive bottom-up merging approach by Liu and Sun. Additionally, we show significant variations in performance between different implementations the same algorithms highlighting the importance of low-level implementation details. Finally, we note that existing parallel min-cut/max-flow algorithms can significantly outperform serial algorithms on large problems but suffers from added overhead on small to medium problems. Implementations of all algorithms are available at https://github.com/patmjen/maxflow_algorithms
Image-Based Alignment of 3D Scans
Sep 14, 2021

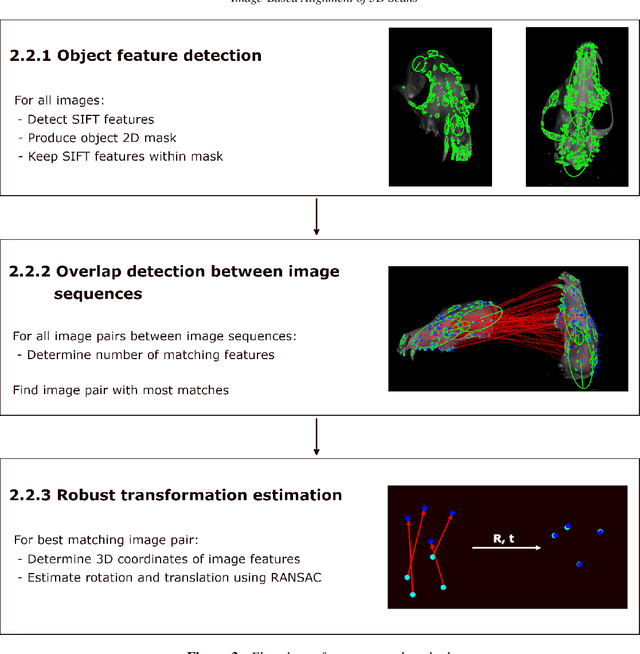

Abstract:Full 3D scanning can efficiently be obtained using structured light scanning combined with a rotation stage. In this setting it is, however, necessary to reposition the object and scan it in different poses in order to cover the entire object. In this case, correspondence between the scans is lost, since the object was moved. In this paper, we propose a fully automatic method for aligning the scans of an object in two different poses. This is done by matching 2D features between images from two poses and utilizing correspondence between the images and the scanned point clouds. To demonstrate the approach, we present the results of scanning three dissimilar objects.
Shape from Projections via Differentiable Forward Projector
Jun 29, 2020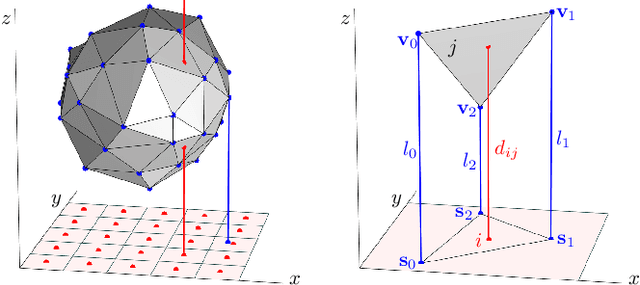


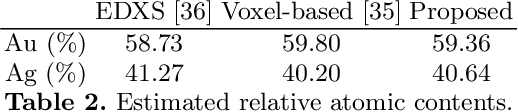
Abstract:In tomography, forward projection of 3D meshes has been mostly studied to simulate data acquisition. However, such works did not consider an inverse process of estimating shapes from projections. In this paper, we propose a differentiable forward projector for 3D meshes, to bridge the gap between the forward model for 3D surfaces and optimization. We view the forward projection as a rendering process, and make it differentiable by extending a recent work in differentiable rasterization. We use the proposed forward projector to reconstruct 3D shapes directly from projections. Experimental results for single-object problems show that our method outperforms the traditional voxel-based methods on noisy simulated data. We also apply our method on real data from electron tomography to estimate the shapes of some nanoparticles.
Assessment of algorithms for mitosis detection in breast cancer histopathology images
Nov 21, 2014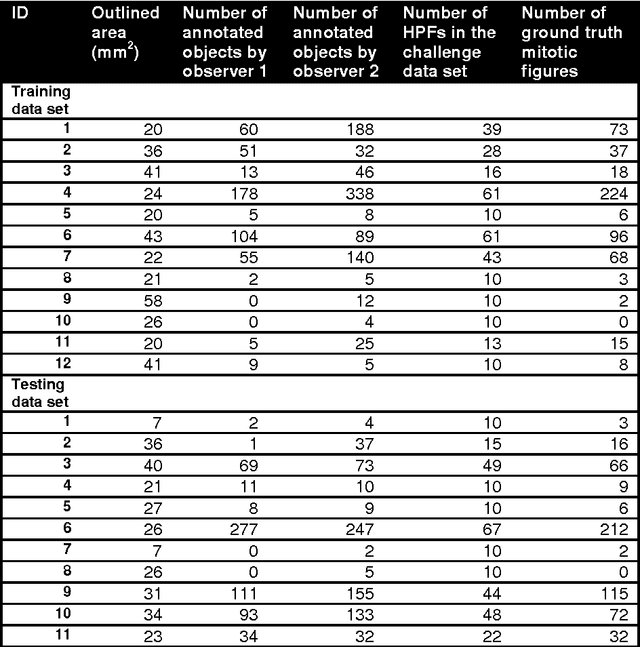
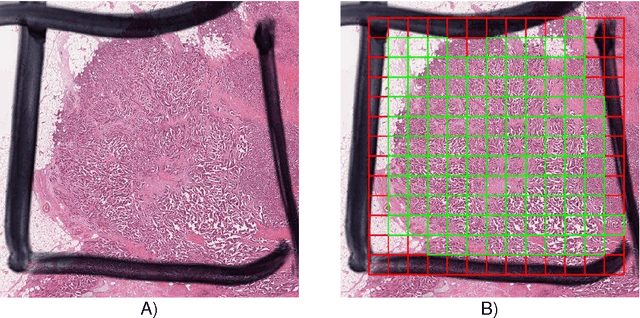
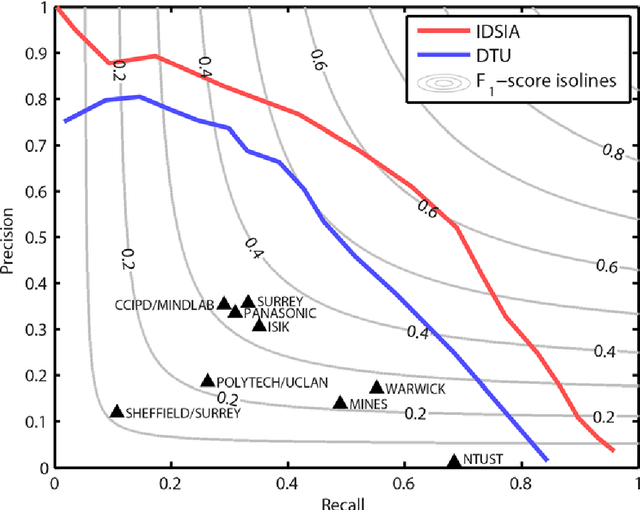
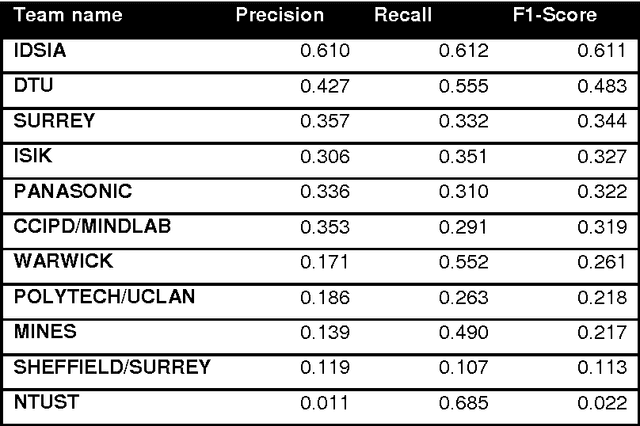
Abstract:The proliferative activity of breast tumors, which is routinely estimated by counting of mitotic figures in hematoxylin and eosin stained histology sections, is considered to be one of the most important prognostic markers. However, mitosis counting is laborious, subjective and may suffer from low inter-observer agreement. With the wider acceptance of whole slide images in pathology labs, automatic image analysis has been proposed as a potential solution for these issues. In this paper, the results from the Assessment of Mitosis Detection Algorithms 2013 (AMIDA13) challenge are described. The challenge was based on a data set consisting of 12 training and 11 testing subjects, with more than one thousand annotated mitotic figures by multiple observers. Short descriptions and results from the evaluation of eleven methods are presented. The top performing method has an error rate that is comparable to the inter-observer agreement among pathologists.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge