Zhanghexuan Ji
A Continual Learning-driven Model for Accurate and Generalizable Segmentation of Clinically Comprehensive and Fine-grained Whole-body Anatomies in CT
Mar 16, 2025Abstract:Precision medicine in the quantitative management of chronic diseases and oncology would be greatly improved if the Computed Tomography (CT) scan of any patient could be segmented, parsed and analyzed in a precise and detailed way. However, there is no such fully annotated CT dataset with all anatomies delineated for training because of the exceptionally high manual cost, the need for specialized clinical expertise, and the time required to finish the task. To this end, we proposed a novel continual learning-driven CT model that can segment complete anatomies presented using dozens of previously partially labeled datasets, dynamically expanding its capacity to segment new ones without compromising previously learned organ knowledge. Existing multi-dataset approaches are not able to dynamically segment new anatomies without catastrophic forgetting and would encounter optimization difficulty or infeasibility when segmenting hundreds of anatomies across the whole range of body regions. Our single unified CT segmentation model, CL-Net, can highly accurately segment a clinically comprehensive set of 235 fine-grained whole-body anatomies. Composed of a universal encoder, multiple optimized and pruned decoders, CL-Net is developed using 13,952 CT scans from 20 public and 16 private high-quality partially labeled CT datasets of various vendors, different contrast phases, and pathologies. Extensive evaluation demonstrates that CL-Net consistently outperforms the upper limit of an ensemble of 36 specialist nnUNets trained per dataset with the complexity of 5% model size and significantly surpasses the segmentation accuracy of recent leading Segment Anything-style medical image foundation models by large margins. Our continual learning-driven CL-Net model would lay a solid foundation to facilitate many downstream tasks of oncology and chronic diseases using the most widely adopted CT imaging.
From Slices to Sequences: Autoregressive Tracking Transformer for Cohesive and Consistent 3D Lymph Node Detection in CT Scans
Mar 11, 2025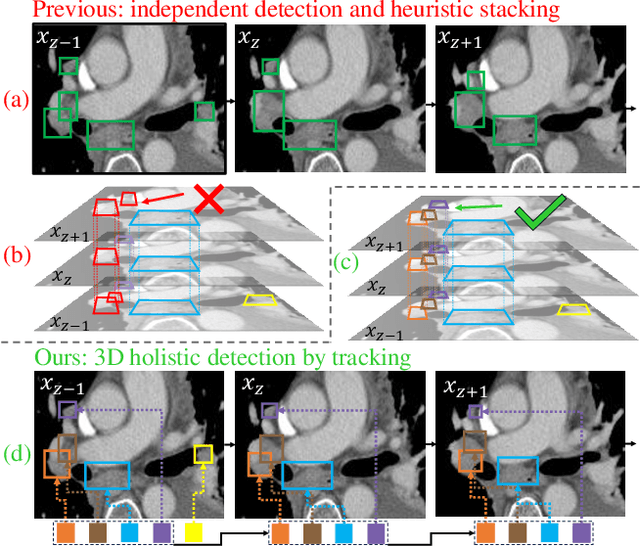

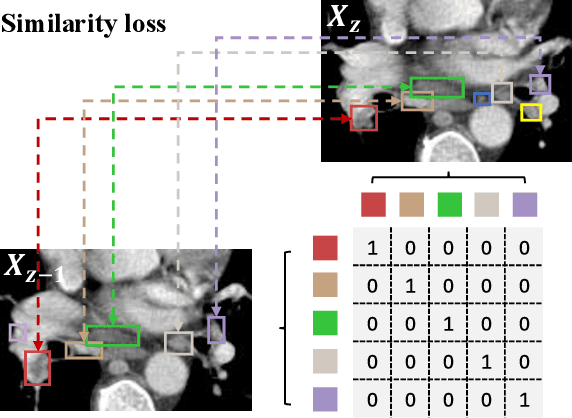
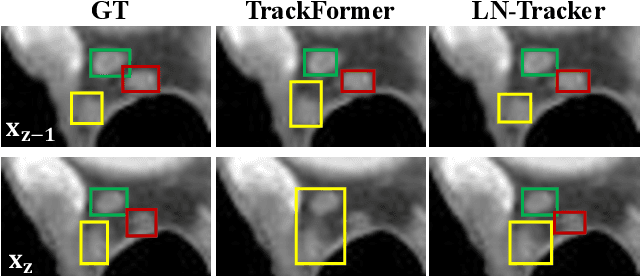
Abstract:Lymph node (LN) assessment is an essential task in the routine radiology workflow, providing valuable insights for cancer staging, treatment planning and beyond. Identifying scatteredly-distributed and low-contrast LNs in 3D CT scans is highly challenging, even for experienced clinicians. Previous lesion and LN detection methods demonstrate effectiveness of 2.5D approaches (i.e, using 2D network with multi-slice inputs), leveraging pretrained 2D model weights and showing improved accuracy as compared to separate 2D or 3D detectors. However, slice-based 2.5D detectors do not explicitly model inter-slice consistency for LN as a 3D object, requiring heuristic post-merging steps to generate final 3D LN instances, which can involve tuning a set of parameters for each dataset. In this work, we formulate 3D LN detection as a tracking task and propose LN-Tracker, a novel LN tracking transformer, for joint end-to-end detection and 3D instance association. Built upon DETR-based detector, LN-Tracker decouples transformer decoder's query into the track and detection groups, where the track query autoregressively follows previously tracked LN instances along the z-axis of a CT scan. We design a new transformer decoder with masked attention module to align track query's content to the context of current slice, meanwhile preserving detection query's high accuracy in current slice. An inter-slice similarity loss is introduced to encourage cohesive LN association between slices. Extensive evaluation on four lymph node datasets shows LN-Tracker's superior performance, with at least 2.7% gain in average sensitivity when compared to other top 3D/2.5D detectors. Further validation on public lung nodule and prostate tumor detection tasks confirms the generalizability of LN-Tracker as it achieves top performance on both tasks. Datasets will be released upon acceptance.
Low-Rank Continual Pyramid Vision Transformer: Incrementally Segment Whole-Body Organs in CT with Light-Weighted Adaptation
Oct 07, 2024Abstract:Deep segmentation networks achieve high performance when trained on specific datasets. However, in clinical practice, it is often desirable that pretrained segmentation models can be dynamically extended to enable segmenting new organs without access to previous training datasets or without training from scratch. This would ensure a much more efficient model development and deployment paradigm accounting for the patient privacy and data storage issues. This clinically preferred process can be viewed as a continual semantic segmentation (CSS) problem. Previous CSS works would either experience catastrophic forgetting or lead to unaffordable memory costs as models expand. In this work, we propose a new continual whole-body organ segmentation model with light-weighted low-rank adaptation (LoRA). We first train and freeze a pyramid vision transformer (PVT) base segmentation model on the initial task, then continually add light-weighted trainable LoRA parameters to the frozen model for each new learning task. Through a holistically exploration of the architecture modification, we identify three most important layers (i.e., patch-embedding, multi-head attention and feed forward layers) that are critical in adapting to the new segmentation tasks, while retaining the majority of the pretrained parameters fixed. Our proposed model continually segments new organs without catastrophic forgetting and meanwhile maintaining a low parameter increasing rate. Continually trained and tested on four datasets covering different body parts of a total of 121 organs, results show that our model achieves high segmentation accuracy, closely reaching the PVT and nnUNet upper bounds, and significantly outperforms other regularization-based CSS methods. When comparing to the leading architecture-based CSS method, our model has a substantial lower parameter increasing rate while achieving comparable performance.
Continual Domain Adversarial Adaptation via Double-Head Discriminators
Feb 05, 2024Abstract:Domain adversarial adaptation in a continual setting poses a significant challenge due to the limitations on accessing previous source domain data. Despite extensive research in continual learning, the task of adversarial adaptation cannot be effectively accomplished using only a small number of stored source domain data, which is a standard setting in memory replay approaches. This limitation arises from the erroneous empirical estimation of $\gH$-divergence with few source domain samples. To tackle this problem, we propose a double-head discriminator algorithm, by introducing an addition source-only domain discriminator that are trained solely on source learning phase. We prove that with the introduction of a pre-trained source-only domain discriminator, the empirical estimation error of $\gH$-divergence related adversarial loss is reduced from the source domain side. Further experiments on existing domain adaptation benchmark show that our proposed algorithm achieves more than 2$\%$ improvement on all categories of target domain adaptation task while significantly mitigating the forgetting on source domain.
Continual Segment: Towards a Single, Unified and Accessible Continual Segmentation Model of 143 Whole-body Organs in CT Scans
Feb 04, 2023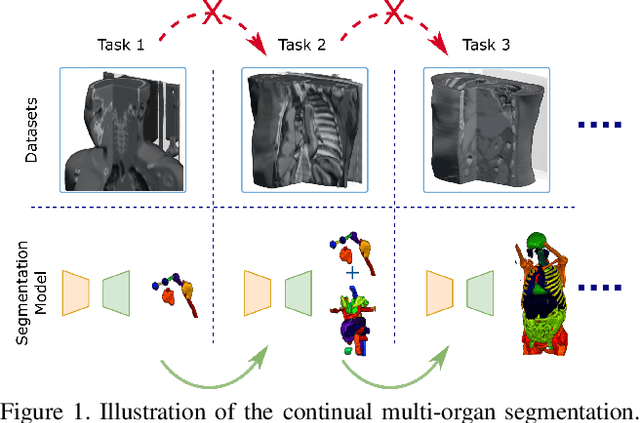
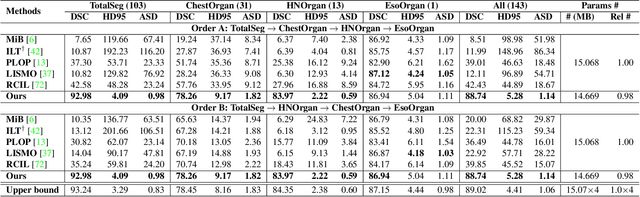
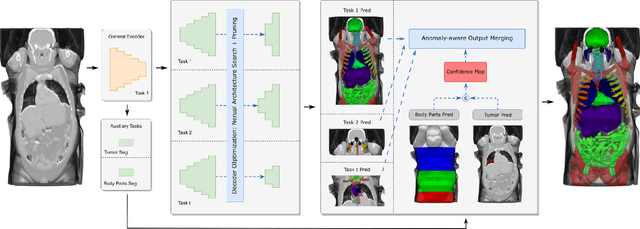
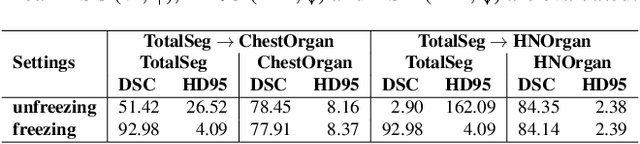
Abstract:Deep learning empowers the mainstream medical image segmentation methods. Nevertheless current deep segmentation approaches are not capable of efficiently and effectively adapting and updating the trained models when new incremental segmentation classes (along with new training datasets or not) are required to be added. In real clinical environment, it can be preferred that segmentation models could be dynamically extended to segment new organs/tumors without the (re-)access to previous training datasets due to obstacles of patient privacy and data storage. This process can be viewed as a continual semantic segmentation (CSS) problem, being understudied for multi-organ segmentation. In this work, we propose a new architectural CSS learning framework to learn a single deep segmentation model for segmenting a total of 143 whole-body organs. Using the encoder/decoder network structure, we demonstrate that a continually-trained then frozen encoder coupled with incrementally-added decoders can extract and preserve sufficiently representative image features for new classes to be subsequently and validly segmented. To maintain a single network model complexity, we trim each decoder progressively using neural architecture search and teacher-student based knowledge distillation. To incorporate with both healthy and pathological organs appearing in different datasets, a novel anomaly-aware and confidence learning module is proposed to merge the overlapped organ predictions, originated from different decoders. Trained and validated on 3D CT scans of 2500+ patients from four datasets, our single network can segment total 143 whole-body organs with very high accuracy, closely reaching the upper bound performance level by training four separate segmentation models (i.e., one model per dataset/task).
Progressive Voronoi Diagram Subdivision: Towards A Holistic Geometric Framework for Exemplar-free Class-Incremental Learning
Jul 28, 2022



Abstract:Exemplar-free Class-incremental Learning (CIL) is a challenging problem because rehearsing data from previous phases is strictly prohibited, causing catastrophic forgetting of Deep Neural Networks (DNNs). In this paper, we present iVoro, a holistic framework for CIL, derived from computational geometry. We found Voronoi Diagram (VD), a classical model for space subdivision, is especially powerful for solving the CIL problem, because VD itself can be constructed favorably in an incremental manner -- the newly added sites (classes) will only affect the proximate classes, making the non-contiguous classes hardly forgettable. Further, in order to find a better set of centers for VD construction, we colligate DNN with VD using Power Diagram and show that the VD structure can be optimized by integrating local DNN models using a divide-and-conquer algorithm. Moreover, our VD construction is not restricted to the deep feature space, but is also applicable to multiple intermediate feature spaces, promoting VD to be multi-centered VD (CIVD) that efficiently captures multi-grained features from DNN. Importantly, iVoro is also capable of handling uncertainty-aware test-time Voronoi cell assignment and has exhibited high correlations between geometric uncertainty and predictive accuracy (up to ~0.9). Putting everything together, iVoro achieves up to 25.26%, 37.09%, and 33.21% improvements on CIFAR-100, TinyImageNet, and ImageNet-Subset, respectively, compared to the state-of-the-art non-exemplar CIL approaches. In conclusion, iVoro enables highly accurate, privacy-preserving, and geometrically interpretable CIL that is particularly useful when cross-phase data sharing is forbidden, e.g. in medical applications. Our code is available at https://machunwei.github.io/ivoro.
A Bayesian Detect to Track System for Robust Visual Object Tracking and Semi-Supervised Model Learning
May 05, 2022



Abstract:Object tracking is one of the fundamental problems in visual recognition tasks and has achieved significant improvements in recent years. The achievements often come with the price of enormous hardware consumption and expensive labor effort for consecutive labeling. A missing ingredient for robust tracking is achieving performance with minimal modification on network structure and semi-supervised learning intermittent labeled frames. In this paper, we ad-dress these problems in a Bayesian tracking and detection framework parameterized by neural network outputs. In our framework, the tracking and detection process is formulated in a probabilistic way as multi-objects dynamics and network detection uncertainties. With our formulation, we propose a particle filter-based approximate sampling algorithm for tracking object state estimation. Based on our particle filter inference algorithm, a semi-supervised learn-ing algorithm is utilized for learning tracking network on intermittent labeled frames by variational inference. In our experiments, we provide both mAP and probability-based detection measurements for comparison between our algorithm with non-Bayesian solutions. We also train a semi-supervised tracking network on M2Cai16-Tool-Locations Dataset and compare our results with supervised learning on fully labeled frames.
FedMM: Saddle Point Optimization for Federated Adversarial Domain Adaptation
Oct 24, 2021



Abstract:Federated adversary domain adaptation is a unique distributed minimax training task due to the prevalence of label imbalance among clients, with each client only seeing a subset of the classes of labels required to train a global model. To tackle this problem, we propose a distributed minimax optimizer referred to as FedMM, designed specifically for the federated adversary domain adaptation problem. It works well even in the extreme case where each client has different label classes and some clients only have unsupervised tasks. We prove that FedMM ensures convergence to a stationary point with domain-shifted unsupervised data. On a variety of benchmark datasets, extensive experiments show that FedMM consistently achieves either significant communication savings or significant accuracy improvements over federated optimizers based on the gradient descent ascent (GDA) algorithm. When training from scratch, for example, it outperforms other GDA based federated average methods by around $20\%$ in accuracy over the same communication rounds; and it consistently outperforms when training from pre-trained models with an accuracy improvement from $5.4\%$ to $9\%$ for different networks.
Improving Joint Learning of Chest X-Ray and Radiology Report by Word Region Alignment
Sep 04, 2021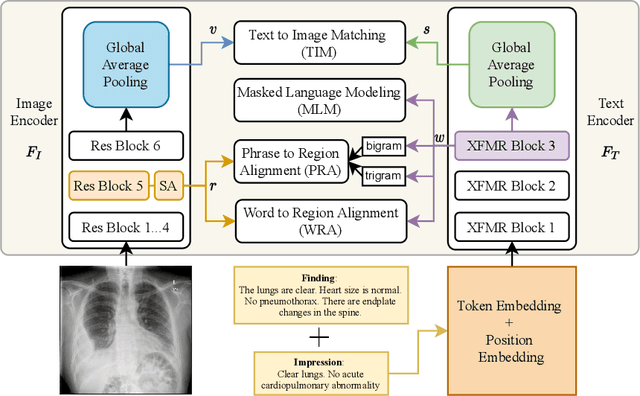
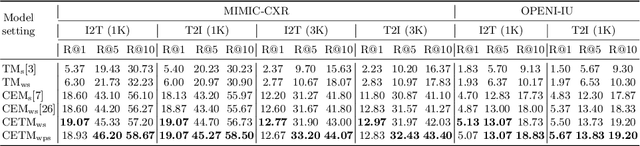
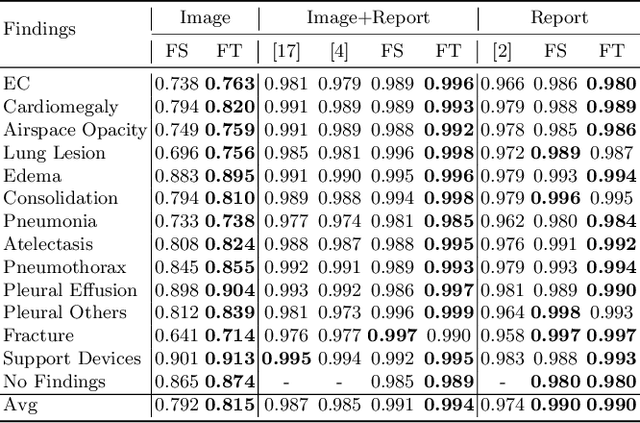
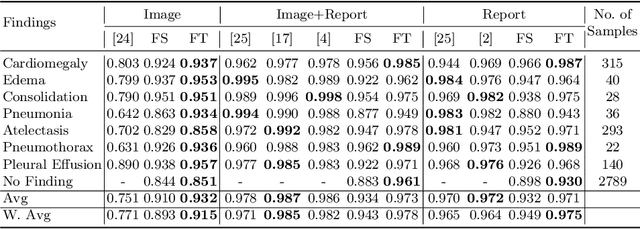
Abstract:Self-supervised learning provides an opportunity to explore unlabeled chest X-rays and their associated free-text reports accumulated in clinical routine without manual supervision. This paper proposes a Joint Image Text Representation Learning Network (JoImTeRNet) for pre-training on chest X-ray images and their radiology reports. The model was pre-trained on both the global image-sentence level and the local image region-word level for visual-textual matching. Both are bidirectionally constrained on Cross-Entropy based and ranking-based Triplet Matching Losses. The region-word matching is calculated using the attention mechanism without direct supervision about their mapping. The pre-trained multi-modal representation learning paves the way for downstream tasks concerning image and/or text encoding. We demonstrate the representation learning quality by cross-modality retrievals and multi-label classifications on two datasets: OpenI-IU and MIMIC-CXR
LAViTeR: Learning Aligned Visual and Textual Representations Assisted by Image and Caption Generation
Sep 04, 2021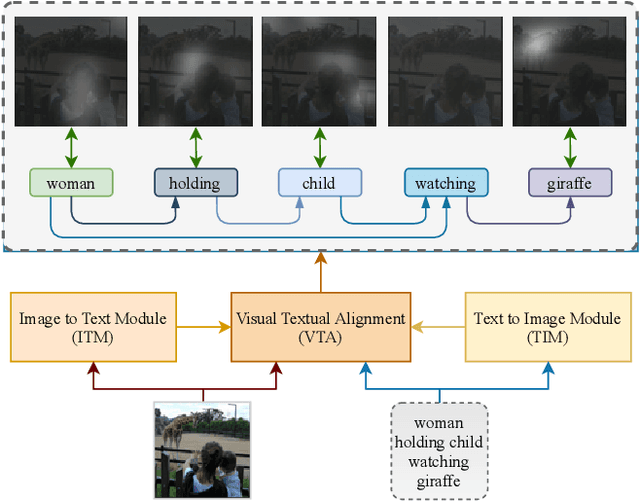
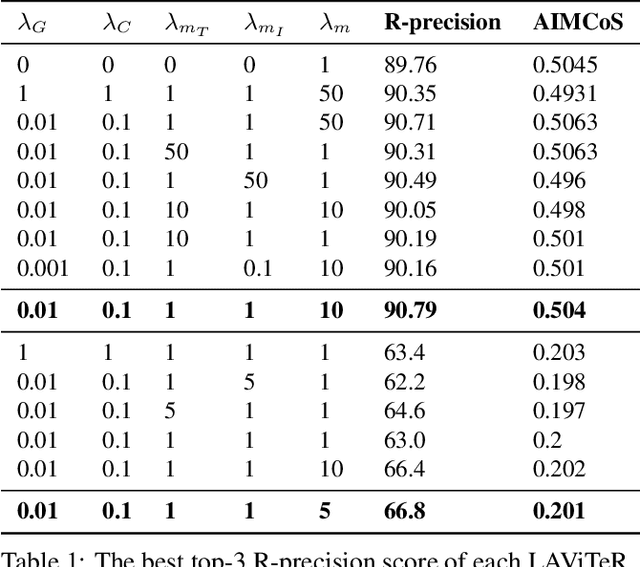
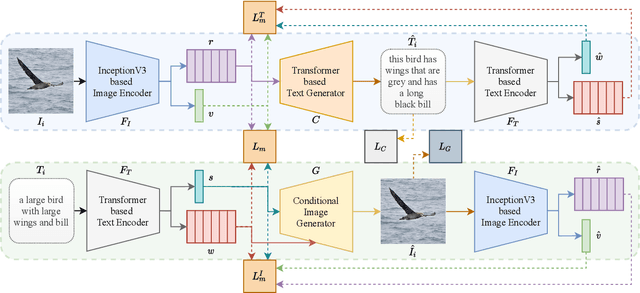
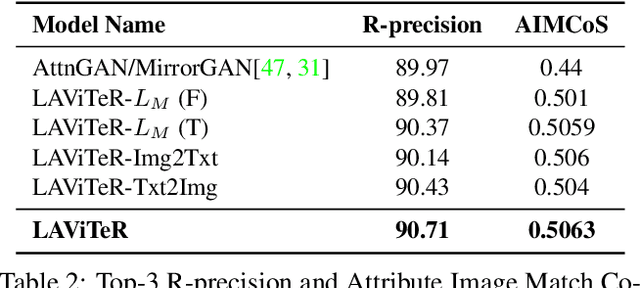
Abstract:Pre-training visual and textual representations from large-scale image-text pairs is becoming a standard approach for many downstream vision-language tasks. The transformer-based models learn inter and intra-modal attention through a list of self-supervised learning tasks. This paper proposes LAViTeR, a novel architecture for visual and textual representation learning. The main module, Visual Textual Alignment (VTA) will be assisted by two auxiliary tasks, GAN-based image synthesis and Image Captioning. We also propose a new evaluation metric measuring the similarity between the learnt visual and textual embedding. The experimental results on two public datasets, CUB and MS-COCO, demonstrate superior visual and textual representation alignment in the joint feature embedding space
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge