Qifeng Wang
An Adaptive ICP LiDAR Odometry Based on Reliable Initial Pose
Sep 26, 2025Abstract:As a key technology for autonomous navigation and positioning in mobile robots, light detection and ranging (LiDAR) odometry is widely used in autonomous driving applications. The Iterative Closest Point (ICP)-based methods have become the core technique in LiDAR odometry due to their efficient and accurate point cloud registration capability. However, some existing ICP-based methods do not consider the reliability of the initial pose, which may cause the method to converge to a local optimum. Furthermore, the absence of an adaptive mechanism hinders the effective handling of complex dynamic environments, resulting in a significant degradation of registration accuracy. To address these issues, this paper proposes an adaptive ICP-based LiDAR odometry method that relies on a reliable initial pose. First, distributed coarse registration based on density filtering is employed to obtain the initial pose estimation. The reliable initial pose is then selected by comparing it with the motion prediction pose, reducing the initial error between the source and target point clouds. Subsequently, by combining the current and historical errors, the adaptive threshold is dynamically adjusted to accommodate the real-time changes in the dynamic environment. Finally, based on the reliable initial pose and the adaptive threshold, point-to-plane adaptive ICP registration is performed from the current frame to the local map, achieving high-precision alignment of the source and target point clouds. Extensive experiments on the public KITTI dataset demonstrate that the proposed method outperforms existing approaches and significantly enhances the accuracy of LiDAR odometry.
A Continual Learning-driven Model for Accurate and Generalizable Segmentation of Clinically Comprehensive and Fine-grained Whole-body Anatomies in CT
Mar 16, 2025Abstract:Precision medicine in the quantitative management of chronic diseases and oncology would be greatly improved if the Computed Tomography (CT) scan of any patient could be segmented, parsed and analyzed in a precise and detailed way. However, there is no such fully annotated CT dataset with all anatomies delineated for training because of the exceptionally high manual cost, the need for specialized clinical expertise, and the time required to finish the task. To this end, we proposed a novel continual learning-driven CT model that can segment complete anatomies presented using dozens of previously partially labeled datasets, dynamically expanding its capacity to segment new ones without compromising previously learned organ knowledge. Existing multi-dataset approaches are not able to dynamically segment new anatomies without catastrophic forgetting and would encounter optimization difficulty or infeasibility when segmenting hundreds of anatomies across the whole range of body regions. Our single unified CT segmentation model, CL-Net, can highly accurately segment a clinically comprehensive set of 235 fine-grained whole-body anatomies. Composed of a universal encoder, multiple optimized and pruned decoders, CL-Net is developed using 13,952 CT scans from 20 public and 16 private high-quality partially labeled CT datasets of various vendors, different contrast phases, and pathologies. Extensive evaluation demonstrates that CL-Net consistently outperforms the upper limit of an ensemble of 36 specialist nnUNets trained per dataset with the complexity of 5% model size and significantly surpasses the segmentation accuracy of recent leading Segment Anything-style medical image foundation models by large margins. Our continual learning-driven CL-Net model would lay a solid foundation to facilitate many downstream tasks of oncology and chronic diseases using the most widely adopted CT imaging.
Multi-modal Intermediate Feature Interaction AutoEncoder for Overall Survival Prediction of Esophageal Squamous Cell Cancer
Aug 23, 2024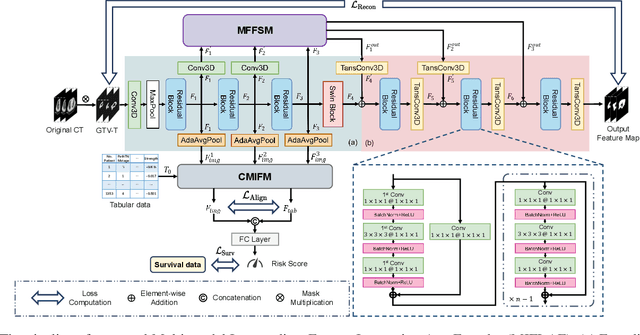
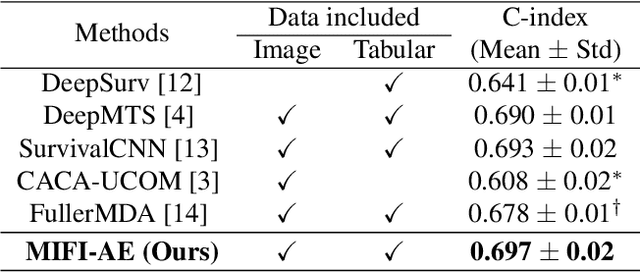
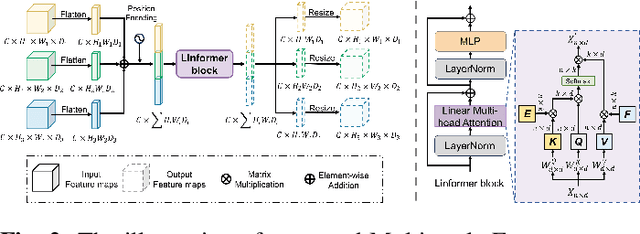
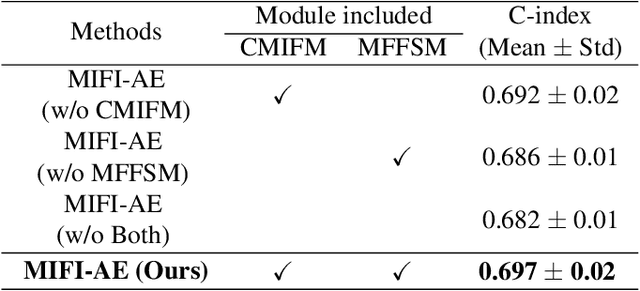
Abstract:Survival prediction for esophageal squamous cell cancer (ESCC) is crucial for doctors to assess a patient's condition and tailor treatment plans. The application and development of multi-modal deep learning in this field have attracted attention in recent years. However, the prognostically relevant features between cross-modalities have not been further explored in previous studies, which could hinder the performance of the model. Furthermore, the inherent semantic gap between different modal feature representations is also ignored. In this work, we propose a novel autoencoder-based deep learning model to predict the overall survival of the ESCC. Two novel modules were designed for multi-modal prognosis-related feature reinforcement and modeling ability enhancement. In addition, a novel joint loss was proposed to make the multi-modal feature representations more aligned. Comparison and ablation experiments demonstrated that our model can achieve satisfactory results in terms of discriminative ability, risk stratification, and the effectiveness of the proposed modules.
MMFusion: Multi-modality Diffusion Model for Lymph Node Metastasis Diagnosis in Esophageal Cancer
May 16, 2024Abstract:Esophageal cancer is one of the most common types of cancer worldwide and ranks sixth in cancer-related mortality. Accurate computer-assisted diagnosis of cancer progression can help physicians effectively customize personalized treatment plans. Currently, CT-based cancer diagnosis methods have received much attention for their comprehensive ability to examine patients' conditions. However, multi-modal based methods may likely introduce information redundancy, leading to underperformance. In addition, efficient and effective interactions between multi-modal representations need to be further explored, lacking insightful exploration of prognostic correlation in multi-modality features. In this work, we introduce a multi-modal heterogeneous graph-based conditional feature-guided diffusion model for lymph node metastasis diagnosis based on CT images as well as clinical measurements and radiomics data. To explore the intricate relationships between multi-modal features, we construct a heterogeneous graph. Following this, a conditional feature-guided diffusion approach is applied to eliminate information redundancy. Moreover, we propose a masked relational representation learning strategy, aiming to uncover the latent prognostic correlations and priorities of primary tumor and lymph node image representations. Various experimental results validate the effectiveness of our proposed method. The code is available at https://github.com/wuchengyu123/MMFusion.
Effective Lymph Nodes Detection in CT Scans Using Location Debiased Query Selection and Contrastive Query Representation in Transformer
Apr 04, 2024Abstract:Lymph node (LN) assessment is a critical, indispensable yet very challenging task in the routine clinical workflow of radiology and oncology. Accurate LN analysis is essential for cancer diagnosis, staging, and treatment planning. Finding scatteredly distributed, low-contrast clinically relevant LNs in 3D CT is difficult even for experienced physicians under high inter-observer variations. Previous automatic LN detection works typically yield limited recall and high false positives (FPs) due to adjacent anatomies with similar image intensities, shapes, or textures (vessels, muscles, esophagus, etc). In this work, we propose a new LN DEtection TRansformer, named LN-DETR, to achieve more accurate performance. By enhancing the 2D backbone with a multi-scale 2.5D feature fusion to incorporate 3D context explicitly, more importantly, we make two main contributions to improve the representation quality of LN queries. 1) Considering that LN boundaries are often unclear, an IoU prediction head and a location debiased query selection are proposed to select LN queries of higher localization accuracy as the decoder query's initialization. 2) To reduce FPs, query contrastive learning is employed to explicitly reinforce LN queries towards their best-matched ground-truth queries over unmatched query predictions. Trained and tested on 3D CT scans of 1067 patients (with 10,000+ labeled LNs) via combining seven LN datasets from different body parts (neck, chest, and abdomen) and pathologies/cancers, our method significantly improves the performance of previous leading methods by > 4-5% average recall at the same FP rates in both internal and external testing. We further evaluate on the universal lesion detection task using NIH DeepLesion benchmark, and our method achieves the top performance of 88.46% averaged recall across 0.5 to 4 FPs per image, compared with other leading reported results.
Continual Segment: Towards a Single, Unified and Accessible Continual Segmentation Model of 143 Whole-body Organs in CT Scans
Feb 04, 2023
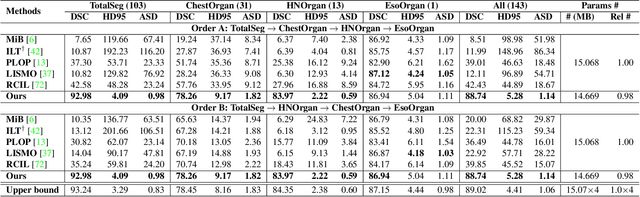
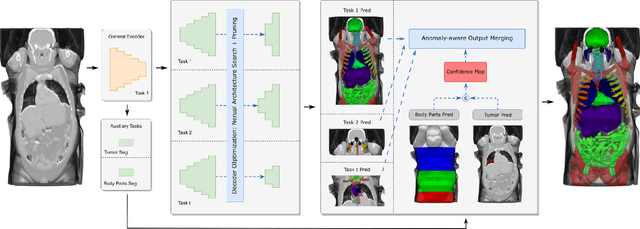
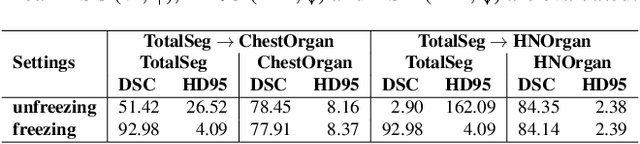
Abstract:Deep learning empowers the mainstream medical image segmentation methods. Nevertheless current deep segmentation approaches are not capable of efficiently and effectively adapting and updating the trained models when new incremental segmentation classes (along with new training datasets or not) are required to be added. In real clinical environment, it can be preferred that segmentation models could be dynamically extended to segment new organs/tumors without the (re-)access to previous training datasets due to obstacles of patient privacy and data storage. This process can be viewed as a continual semantic segmentation (CSS) problem, being understudied for multi-organ segmentation. In this work, we propose a new architectural CSS learning framework to learn a single deep segmentation model for segmenting a total of 143 whole-body organs. Using the encoder/decoder network structure, we demonstrate that a continually-trained then frozen encoder coupled with incrementally-added decoders can extract and preserve sufficiently representative image features for new classes to be subsequently and validly segmented. To maintain a single network model complexity, we trim each decoder progressively using neural architecture search and teacher-student based knowledge distillation. To incorporate with both healthy and pathological organs appearing in different datasets, a novel anomaly-aware and confidence learning module is proposed to merge the overlapped organ predictions, originated from different decoders. Trained and validated on 3D CT scans of 2500+ patients from four datasets, our single network can segment total 143 whole-body organs with very high accuracy, closely reaching the upper bound performance level by training four separate segmentation models (i.e., one model per dataset/task).
Towards a Single Unified Model for Effective Detection, Segmentation, and Diagnosis of Eight Major Cancers Using a Large Collection of CT Scans
Jan 28, 2023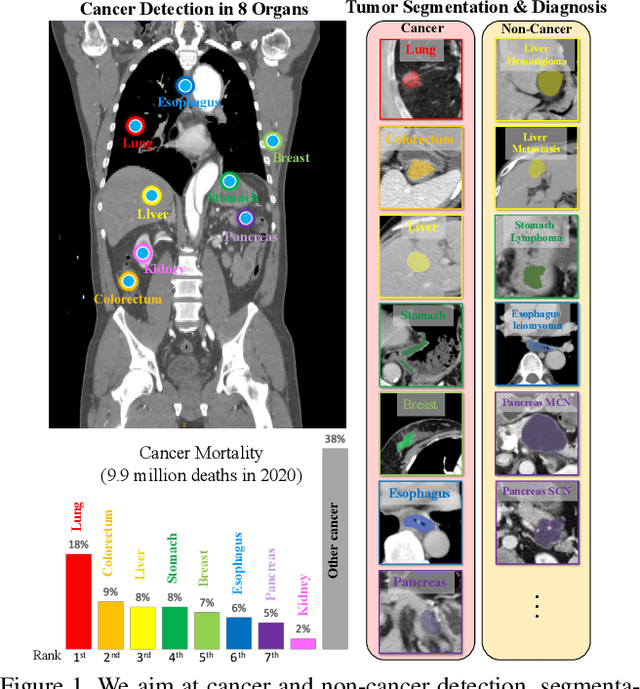
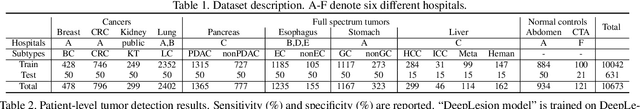
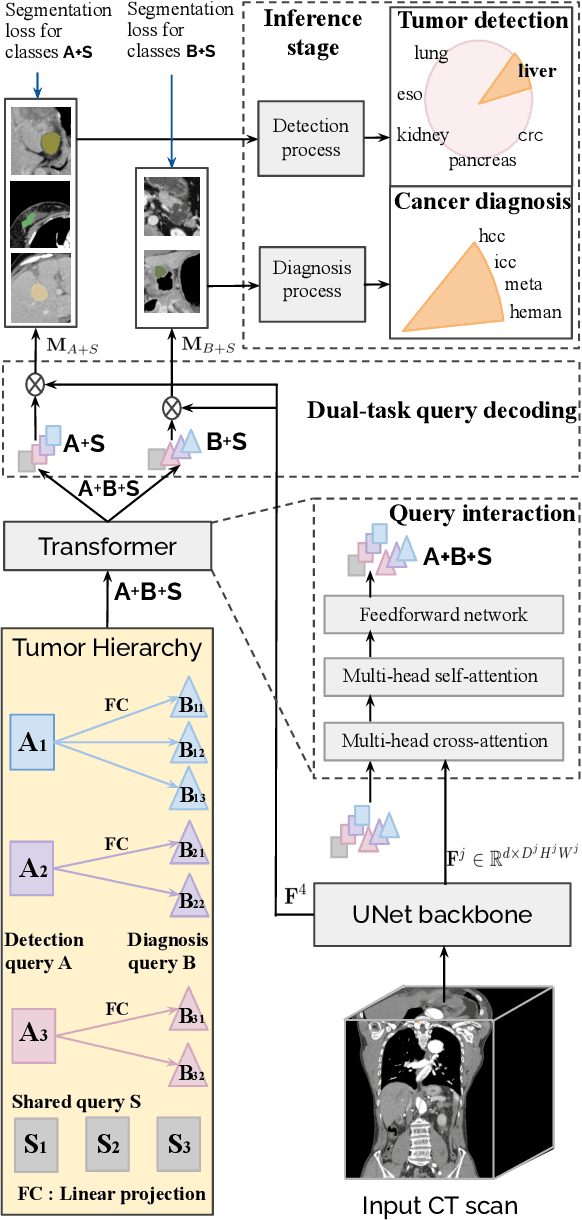
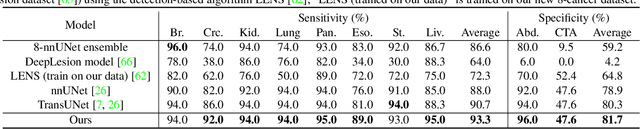
Abstract:Human readers or radiologists routinely perform full-body multi-organ multi-disease detection and diagnosis in clinical practice, while most medical AI systems are built to focus on single organs with a narrow list of a few diseases. This might severely limit AI's clinical adoption. A certain number of AI models need to be assembled non-trivially to match the diagnostic process of a human reading a CT scan. In this paper, we construct a Unified Tumor Transformer (UniT) model to detect (tumor existence and location) and diagnose (tumor characteristics) eight major cancer-prevalent organs in CT scans. UniT is a query-based Mask Transformer model with the output of multi-organ and multi-tumor semantic segmentation. We decouple the object queries into organ queries, detection queries and diagnosis queries, and further establish hierarchical relationships among the three groups. This clinically-inspired architecture effectively assists inter- and intra-organ representation learning of tumors and facilitates the resolution of these complex, anatomically related multi-organ cancer image reading tasks. UniT is trained end-to-end using a curated large-scale CT images of 10,042 patients including eight major types of cancers and occurring non-cancer tumors (all are pathology-confirmed with 3D tumor masks annotated by radiologists). On the test set of 631 patients, UniT has demonstrated strong performance under a set of clinically relevant evaluation metrics, substantially outperforming both multi-organ segmentation methods and an assembly of eight single-organ expert models in tumor detection, segmentation, and diagnosis. Such a unified multi-cancer image reading model (UniT) can significantly reduce the number of false positives produced by combined multi-system models. This moves one step closer towards a universal high-performance cancer screening tool.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge