Mingze Yuan
RODS: Robust Optimization Inspired Diffusion Sampling for Detecting and Reducing Hallucination in Generative Models
Jul 16, 2025Abstract:Diffusion models have achieved state-of-the-art performance in generative modeling, yet their sampling procedures remain vulnerable to hallucinations, often stemming from inaccuracies in score approximation. In this work, we reinterpret diffusion sampling through the lens of optimization and introduce RODS (Robust Optimization-inspired Diffusion Sampler), a novel method that detects and corrects high-risk sampling steps using geometric cues from the loss landscape. RODS enforces smoother sampling trajectories and adaptively adjusts perturbations, reducing hallucinations without retraining and at minimal additional inference cost. Experiments on AFHQv2, FFHQ, and 11k-hands demonstrate that RODS improves both sampling fidelity and robustness, detecting over 70% of hallucinated samples and correcting more than 25%, all while avoiding the introduction of new artifacts.
CycleINR: Cycle Implicit Neural Representation for Arbitrary-Scale Volumetric Super-Resolution of Medical Data
Apr 07, 2024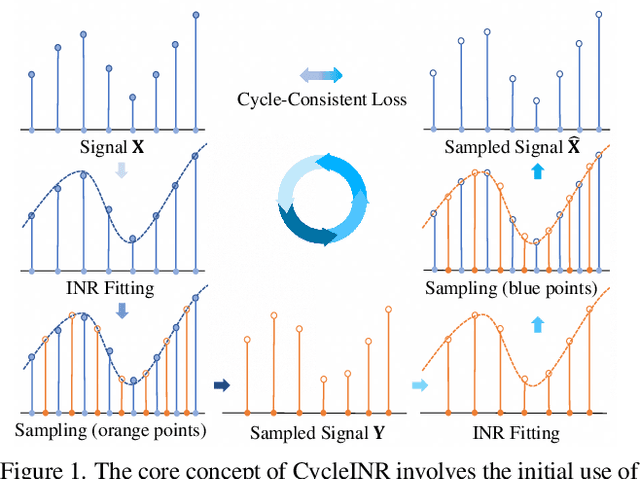
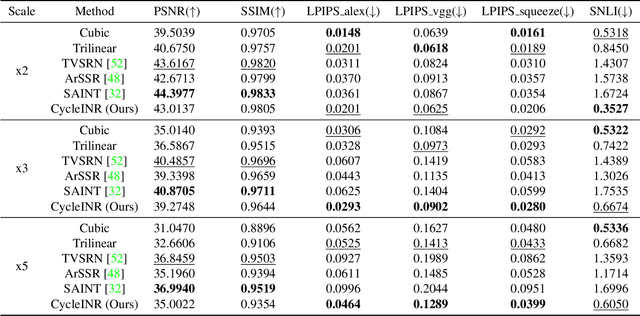
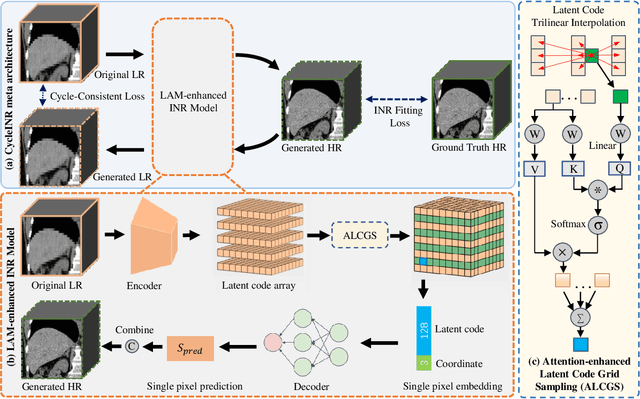
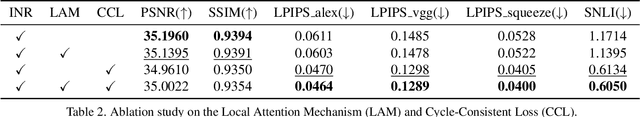
Abstract:In the realm of medical 3D data, such as CT and MRI images, prevalent anisotropic resolution is characterized by high intra-slice but diminished inter-slice resolution. The lowered resolution between adjacent slices poses challenges, hindering optimal viewing experiences and impeding the development of robust downstream analysis algorithms. Various volumetric super-resolution algorithms aim to surmount these challenges, enhancing inter-slice resolution and overall 3D medical imaging quality. However, existing approaches confront inherent challenges: 1) often tailored to specific upsampling factors, lacking flexibility for diverse clinical scenarios; 2) newly generated slices frequently suffer from over-smoothing, degrading fine details, and leading to inter-slice inconsistency. In response, this study presents CycleINR, a novel enhanced Implicit Neural Representation model for 3D medical data volumetric super-resolution. Leveraging the continuity of the learned implicit function, the CycleINR model can achieve results with arbitrary up-sampling rates, eliminating the need for separate training. Additionally, we enhance the grid sampling in CycleINR with a local attention mechanism and mitigate over-smoothing by integrating cycle-consistent loss. We introduce a new metric, Slice-wise Noise Level Inconsistency (SNLI), to quantitatively assess inter-slice noise level inconsistency. The effectiveness of our approach is demonstrated through image quality evaluations on an in-house dataset and a downstream task analysis on the Medical Segmentation Decathlon liver tumor dataset.
Large Language Models Illuminate a Progressive Pathway to Artificial Healthcare Assistant: A Review
Nov 03, 2023



Abstract:With the rapid development of artificial intelligence, large language models (LLMs) have shown promising capabilities in mimicking human-level language comprehension and reasoning. This has sparked significant interest in applying LLMs to enhance various aspects of healthcare, ranging from medical education to clinical decision support. However, medicine involves multifaceted data modalities and nuanced reasoning skills, presenting challenges for integrating LLMs. This paper provides a comprehensive review on the applications and implications of LLMs in medicine. It begins by examining the fundamental applications of general-purpose and specialized LLMs, demonstrating their utilities in knowledge retrieval, research support, clinical workflow automation, and diagnostic assistance. Recognizing the inherent multimodality of medicine, the review then focuses on multimodal LLMs, investigating their ability to process diverse data types like medical imaging and EHRs to augment diagnostic accuracy. To address LLMs' limitations regarding personalization and complex clinical reasoning, the paper explores the emerging development of LLM-powered autonomous agents for healthcare. Furthermore, it summarizes the evaluation methodologies for assessing LLMs' reliability and safety in medical contexts. Overall, this review offers an extensive analysis on the transformative potential of LLMs in modern medicine. It also highlights the pivotal need for continuous optimizations and ethical oversight before these models can be effectively integrated into clinical practice. Visit https://github.com/mingze-yuan/Awesome-LLM-Healthcare for an accompanying GitHub repository containing latest papers.
Improved Prognostic Prediction of Pancreatic Cancer Using Multi-Phase CT by Integrating Neural Distance and Texture-Aware Transformer
Aug 01, 2023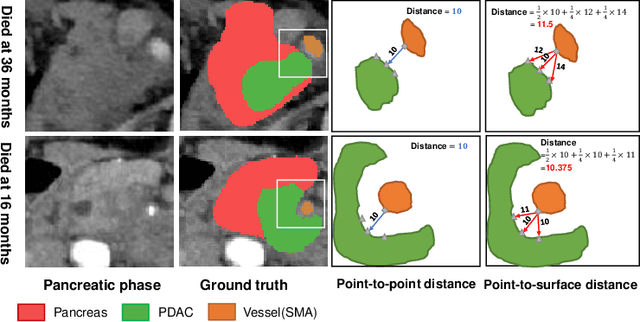
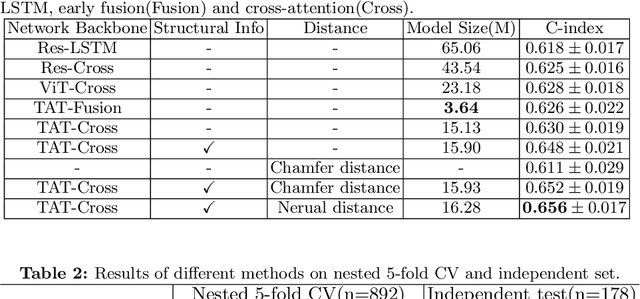
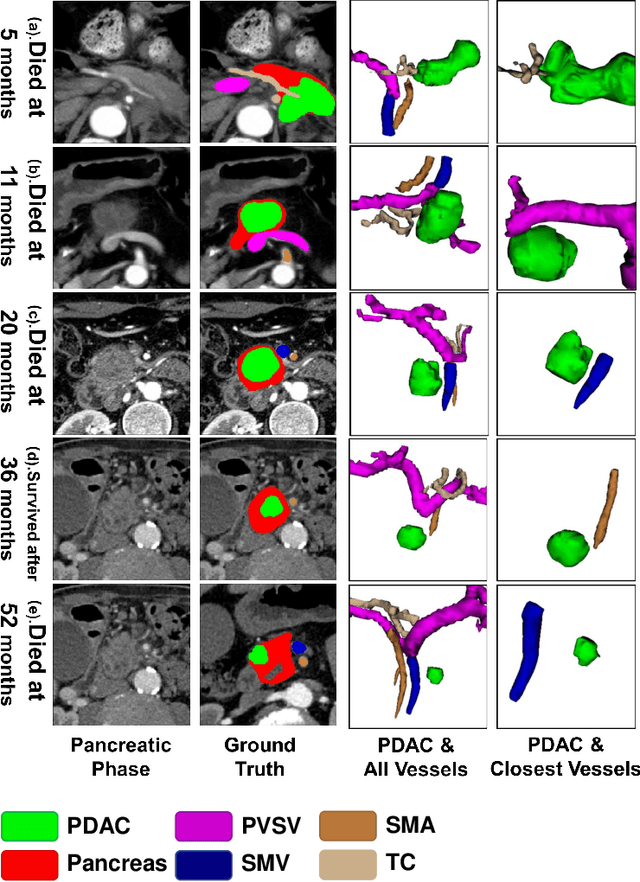
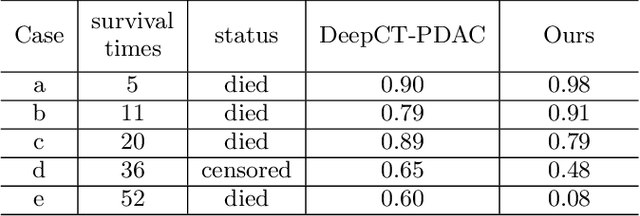
Abstract:Pancreatic ductal adenocarcinoma (PDAC) is a highly lethal cancer in which the tumor-vascular involvement greatly affects the resectability and, thus, overall survival of patients. However, current prognostic prediction methods fail to explicitly and accurately investigate relationships between the tumor and nearby important vessels. This paper proposes a novel learnable neural distance that describes the precise relationship between the tumor and vessels in CT images of different patients, adopting it as a major feature for prognosis prediction. Besides, different from existing models that used CNNs or LSTMs to exploit tumor enhancement patterns on dynamic contrast-enhanced CT imaging, we improved the extraction of dynamic tumor-related texture features in multi-phase contrast-enhanced CT by fusing local and global features using CNN and transformer modules, further enhancing the features extracted across multi-phase CT images. We extensively evaluated and compared the proposed method with existing methods in the multi-center (n=4) dataset with 1,070 patients with PDAC, and statistical analysis confirmed its clinical effectiveness in the external test set consisting of three centers. The developed risk marker was the strongest predictor of overall survival among preoperative factors and it has the potential to be combined with established clinical factors to select patients at higher risk who might benefit from neoadjuvant therapy.
Cluster-Induced Mask Transformers for Effective Opportunistic Gastric Cancer Screening on Non-contrast CT Scans
Jul 16, 2023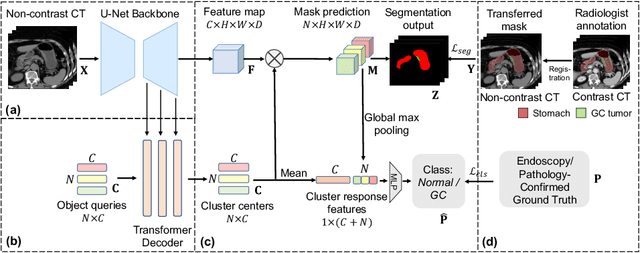
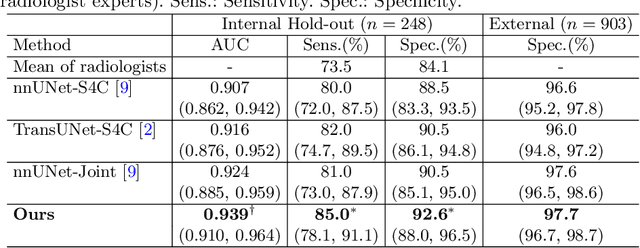
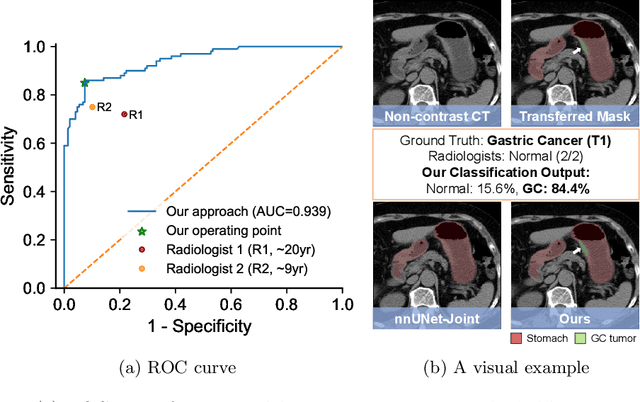
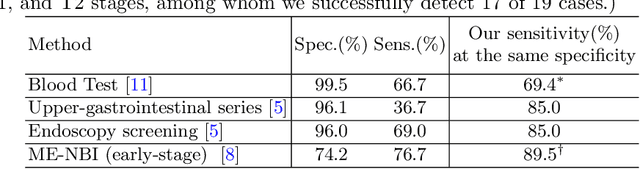
Abstract:Gastric cancer is the third leading cause of cancer-related mortality worldwide, but no guideline-recommended screening test exists. Existing methods can be invasive, expensive, and lack sensitivity to identify early-stage gastric cancer. In this study, we explore the feasibility of using a deep learning approach on non-contrast CT scans for gastric cancer detection. We propose a novel cluster-induced Mask Transformer that jointly segments the tumor and classifies abnormality in a multi-task manner. Our model incorporates learnable clusters that encode the texture and shape prototypes of gastric cancer, utilizing self- and cross-attention to interact with convolutional features. In our experiments, the proposed method achieves a sensitivity of 85.0% and specificity of 92.6% for detecting gastric tumors on a hold-out test set consisting of 100 patients with cancer and 148 normal. In comparison, two radiologists have an average sensitivity of 73.5% and specificity of 84.3%. We also obtain a specificity of 97.7% on an external test set with 903 normal cases. Our approach performs comparably to established state-of-the-art gastric cancer screening tools like blood testing and endoscopy, while also being more sensitive in detecting early-stage cancer. This demonstrates the potential of our approach as a novel, non-invasive, low-cost, and accurate method for opportunistic gastric cancer screening.
Unsupervised Image Denoising with Score Function
Apr 17, 2023Abstract:Though achieving excellent performance in some cases, current unsupervised learning methods for single image denoising usually have constraints in applications. In this paper, we propose a new approach which is more general and applicable to complicated noise models. Utilizing the property of score function, the gradient of logarithmic probability, we define a solving system for denoising. Once the score function of noisy images has been estimated, the denoised result can be obtained through the solving system. Our approach can be applied to multiple noise models, such as the mixture of multiplicative and additive noise combined with structured correlation. Experimental results show that our method is comparable when the noise model is simple, and has good performance in complicated cases where other methods are not applicable or perform poorly.
Devil is in the Queries: Advancing Mask Transformers for Real-world Medical Image Segmentation and Out-of-Distribution Localization
Apr 01, 2023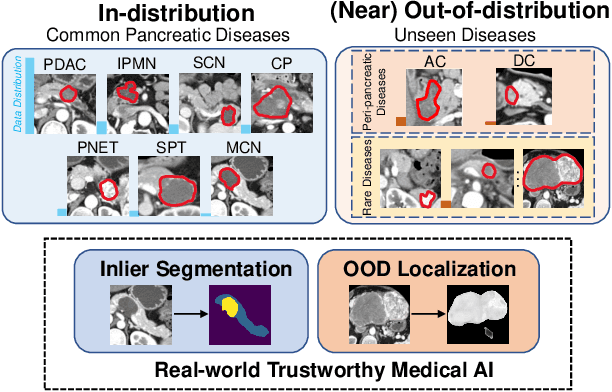
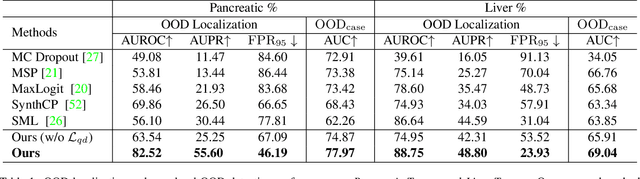
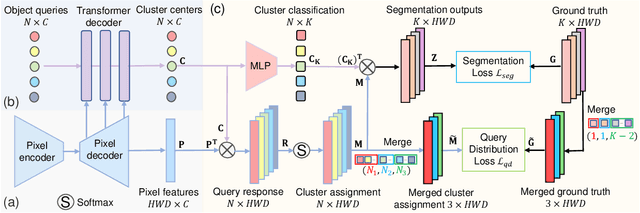
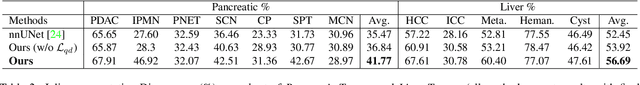
Abstract:Real-world medical image segmentation has tremendous long-tailed complexity of objects, among which tail conditions correlate with relatively rare diseases and are clinically significant. A trustworthy medical AI algorithm should demonstrate its effectiveness on tail conditions to avoid clinically dangerous damage in these out-of-distribution (OOD) cases. In this paper, we adopt the concept of object queries in Mask Transformers to formulate semantic segmentation as a soft cluster assignment. The queries fit the feature-level cluster centers of inliers during training. Therefore, when performing inference on a medical image in real-world scenarios, the similarity between pixels and the queries detects and localizes OOD regions. We term this OOD localization as MaxQuery. Furthermore, the foregrounds of real-world medical images, whether OOD objects or inliers, are lesions. The difference between them is less than that between the foreground and background, possibly misleading the object queries to focus redundantly on the background. Thus, we propose a query-distribution (QD) loss to enforce clear boundaries between segmentation targets and other regions at the query level, improving the inlier segmentation and OOD indication. Our proposed framework is tested on two real-world segmentation tasks, i.e., segmentation of pancreatic and liver tumors, outperforming previous state-of-the-art algorithms by an average of 7.39% on AUROC, 14.69% on AUPR, and 13.79% on FPR95 for OOD localization. On the other hand, our framework improves the performance of inlier segmentation by an average of 5.27% DSC when compared with the leading baseline nnUNet.
Towards a Single Unified Model for Effective Detection, Segmentation, and Diagnosis of Eight Major Cancers Using a Large Collection of CT Scans
Jan 28, 2023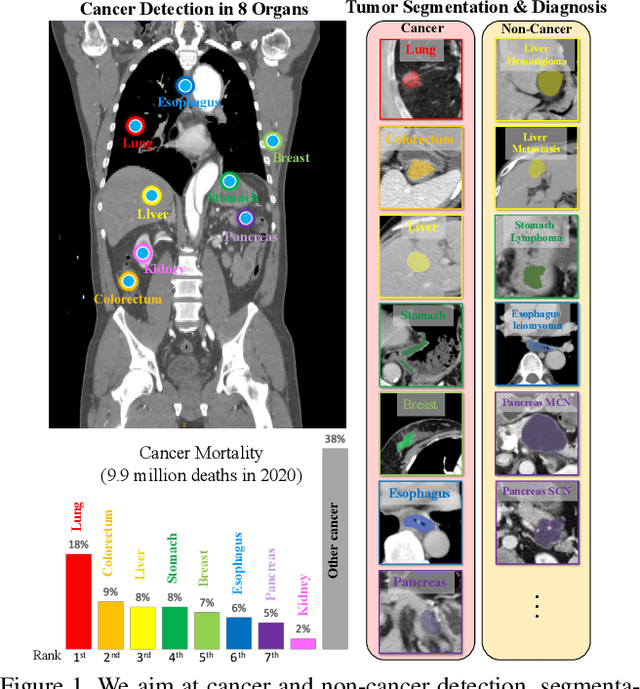
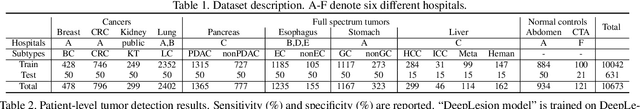
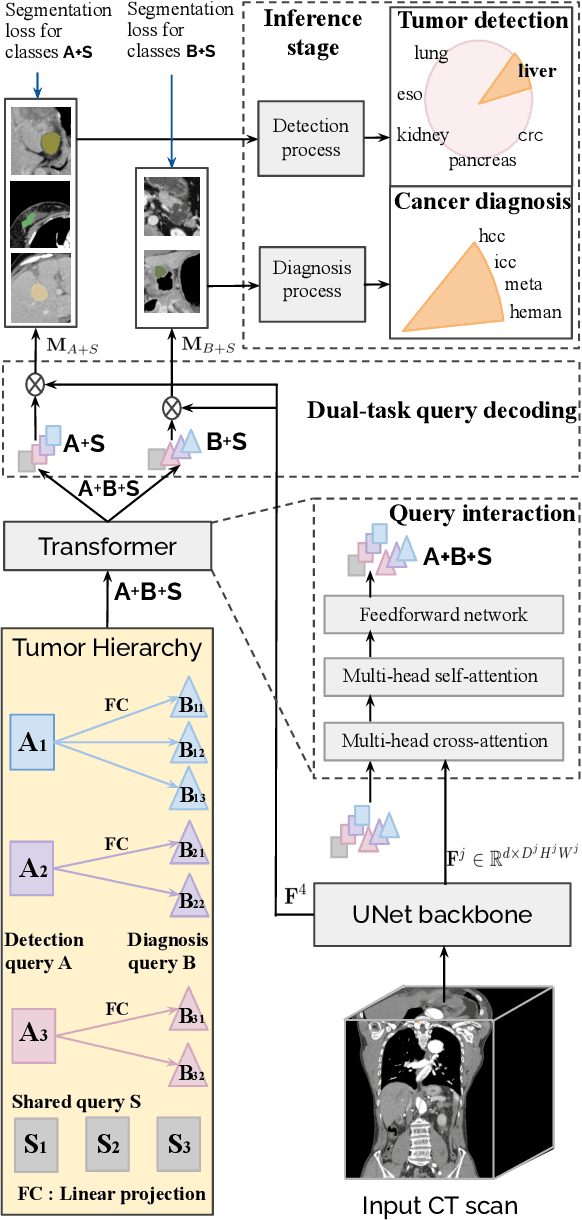
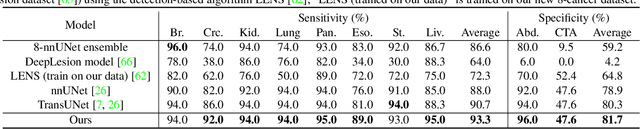
Abstract:Human readers or radiologists routinely perform full-body multi-organ multi-disease detection and diagnosis in clinical practice, while most medical AI systems are built to focus on single organs with a narrow list of a few diseases. This might severely limit AI's clinical adoption. A certain number of AI models need to be assembled non-trivially to match the diagnostic process of a human reading a CT scan. In this paper, we construct a Unified Tumor Transformer (UniT) model to detect (tumor existence and location) and diagnose (tumor characteristics) eight major cancer-prevalent organs in CT scans. UniT is a query-based Mask Transformer model with the output of multi-organ and multi-tumor semantic segmentation. We decouple the object queries into organ queries, detection queries and diagnosis queries, and further establish hierarchical relationships among the three groups. This clinically-inspired architecture effectively assists inter- and intra-organ representation learning of tumors and facilitates the resolution of these complex, anatomically related multi-organ cancer image reading tasks. UniT is trained end-to-end using a curated large-scale CT images of 10,042 patients including eight major types of cancers and occurring non-cancer tumors (all are pathology-confirmed with 3D tumor masks annotated by radiologists). On the test set of 631 patients, UniT has demonstrated strong performance under a set of clinically relevant evaluation metrics, substantially outperforming both multi-organ segmentation methods and an assembly of eight single-organ expert models in tumor detection, segmentation, and diagnosis. Such a unified multi-cancer image reading model (UniT) can significantly reduce the number of false positives produced by combined multi-system models. This moves one step closer towards a universal high-performance cancer screening tool.
Region-Aware Metric Learning for Open World Semantic Segmentation via Meta-Channel Aggregation
May 17, 2022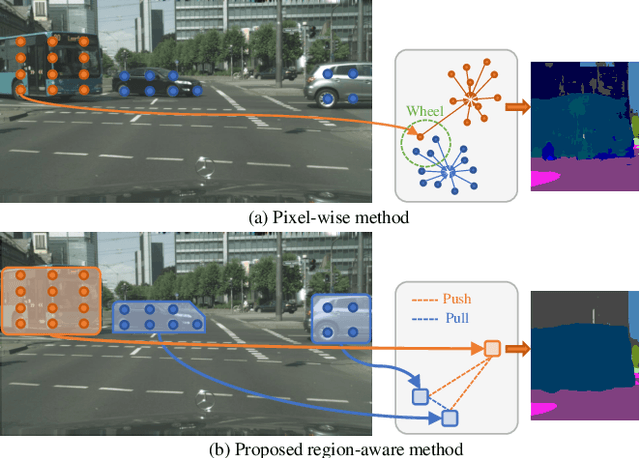
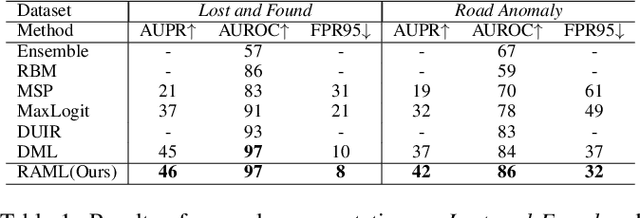
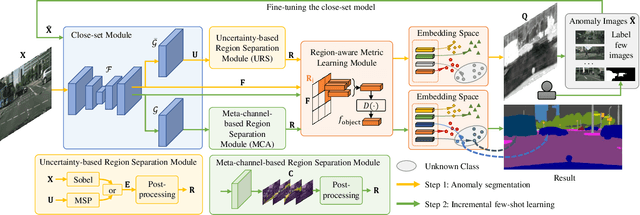
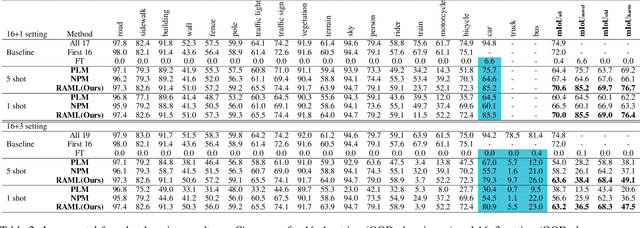
Abstract:As one of the most challenging and practical segmentation tasks, open-world semantic segmentation requires the model to segment the anomaly regions in the images and incrementally learn to segment out-of-distribution (OOD) objects, especially under a few-shot condition. The current state-of-the-art (SOTA) method, Deep Metric Learning Network (DMLNet), relies on pixel-level metric learning, with which the identification of similar regions having different semantics is difficult. Therefore, we propose a method called region-aware metric learning (RAML), which first separates the regions of the images and generates region-aware features for further metric learning. RAML improves the integrity of the segmented anomaly regions. Moreover, we propose a novel meta-channel aggregation (MCA) module to further separate anomaly regions, forming high-quality sub-region candidates and thereby improving the model performance for OOD objects. To evaluate the proposed RAML, we have conducted extensive experiments and ablation studies on Lost And Found and Road Anomaly datasets for anomaly segmentation and the CityScapes dataset for incremental few-shot learning. The results show that the proposed RAML achieves SOTA performance in both stages of open world segmentation. Our code and appendix are available at https://github.com/czifan/RAML.
360° Optical Flow using Tangent Images
Dec 28, 2021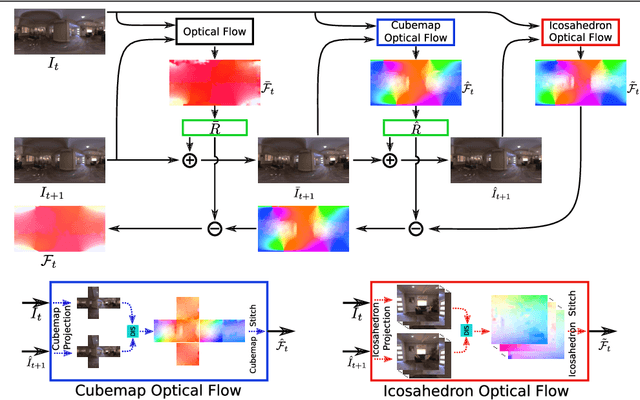
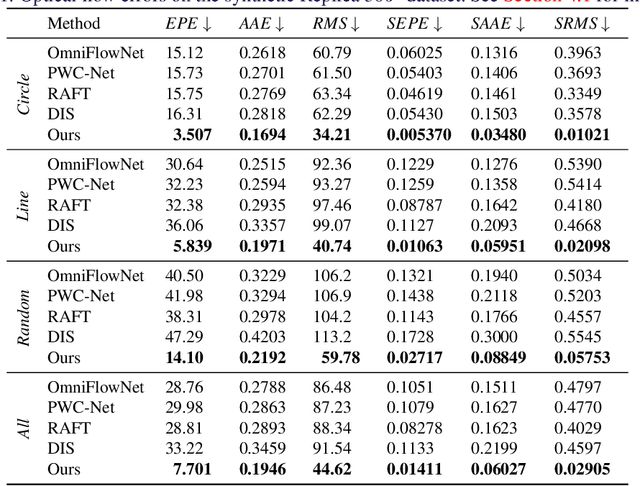

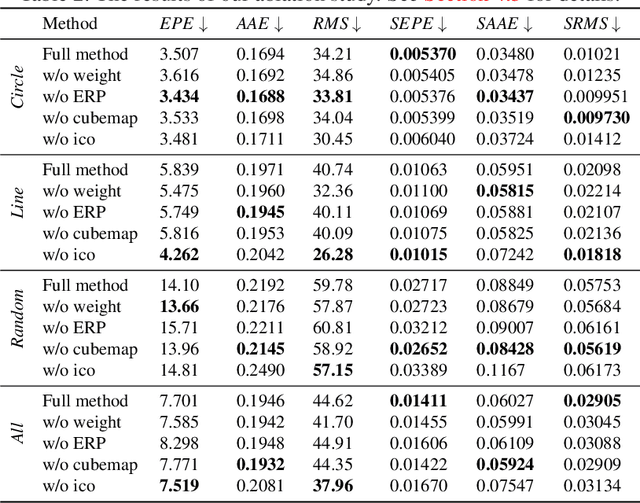
Abstract:Omnidirectional 360{\deg} images have found many promising and exciting applications in computer vision, robotics and other fields, thanks to their increasing affordability, portability and their 360{\deg} field of view. The most common format for storing, processing and visualising 360{\deg} images is equirectangular projection (ERP). However, the distortion introduced by the nonlinear mapping from 360{\deg} image to ERP image is still a barrier that holds back ERP images from being used as easily as conventional perspective images. This is especially relevant when estimating 360{\deg} optical flow, as the distortions need to be mitigated appropriately. In this paper, we propose a 360{\deg} optical flow method based on tangent images. Our method leverages gnomonic projection to locally convert ERP images to perspective images, and uniformly samples the ERP image by projection to a cubemap and regular icosahedron vertices, to incrementally refine the estimated 360{\deg} flow fields even in the presence of large rotations. Our experiments demonstrate the benefits of our proposed method both quantitatively and qualitatively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge