Eran Halperin
Memorize and Rank: Elevating Large Language Models for Clinical Diagnosis Prediction
Jan 28, 2025Abstract:Clinical diagnosis prediction models, when provided with a patient's medical history, aim to detect potential diseases early, facilitating timely intervention and improving prognostic outcomes. However, the inherent scarcity of patient data and large disease candidate space often pose challenges in developing satisfactory models for this intricate task. The exploration of leveraging Large Language Models (LLMs) for encapsulating clinical decision processes has been limited. We introduce MERA, a clinical diagnosis prediction model that bridges pertaining natural language knowledge with medical practice. We apply hierarchical contrastive learning on a disease candidate ranking list to alleviate the large decision space issue. With concept memorization through fine-tuning, we bridge the natural language clinical knowledge with medical codes. Experimental results on MIMIC-III and IV datasets show that MERA achieves the state-of-the-art diagnosis prediction performance and dramatically elevates the diagnosis prediction capabilities of generative LMs.
Guided Discrete Diffusion for Electronic Health Record Generation
Apr 18, 2024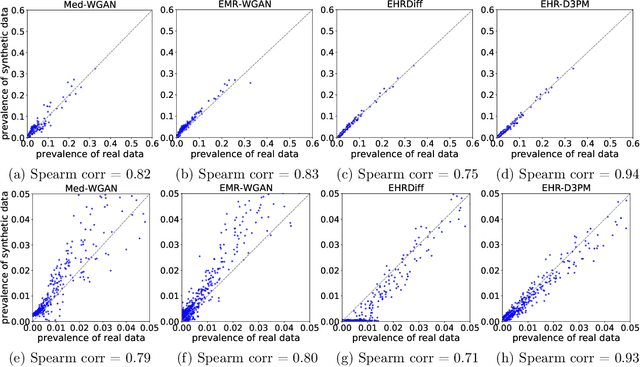

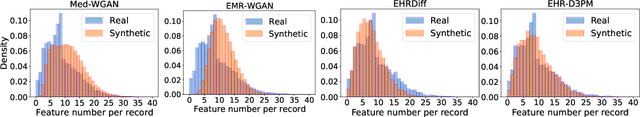

Abstract:Electronic health records (EHRs) are a pivotal data source that enables numerous applications in computational medicine, e.g., disease progression prediction, clinical trial design, and health economics and outcomes research. Despite wide usability, their sensitive nature raises privacy and confidentially concerns, which limit potential use cases. To tackle these challenges, we explore the use of generative models to synthesize artificial, yet realistic EHRs. While diffusion-based methods have recently demonstrated state-of-the-art performance in generating other data modalities and overcome the training instability and mode collapse issues that plague previous GAN-based approaches, their applications in EHR generation remain underexplored. The discrete nature of tabular medical code data in EHRs poses challenges for high-quality data generation, especially for continuous diffusion models. To this end, we introduce a novel tabular EHR generation method, EHR-D3PM, which enables both unconditional and conditional generation using the discrete diffusion model. Our experiments demonstrate that EHR-D3PM significantly outperforms existing generative baselines on comprehensive fidelity and utility metrics while maintaining less membership vulnerability risks. Furthermore, we show EHR-D3PM is effective as a data augmentation method and enhances performance on downstream tasks when combined with real data.
XAIQA: Explainer-Based Data Augmentation for Extractive Question Answering
Dec 06, 2023Abstract:Extractive question answering (QA) systems can enable physicians and researchers to query medical records, a foundational capability for designing clinical studies and understanding patient medical history. However, building these systems typically requires expert-annotated QA pairs. Large language models (LLMs), which can perform extractive QA, depend on high quality data in their prompts, specialized for the application domain. We introduce a novel approach, XAIQA, for generating synthetic QA pairs at scale from data naturally available in electronic health records. Our method uses the idea of a classification model explainer to generate questions and answers about medical concepts corresponding to medical codes. In an expert evaluation with two physicians, our method identifies $2.2\times$ more semantic matches and $3.8\times$ more clinical abbreviations than two popular approaches that use sentence transformers to create QA pairs. In an ML evaluation, adding our QA pairs improves performance of GPT-4 as an extractive QA model, including on difficult questions. In both the expert and ML evaluations, we examine trade-offs between our method and sentence transformers for QA pair generation depending on question difficulty.
Surpassing GPT-4 Medical Coding with a Two-Stage Approach
Nov 22, 2023Abstract:Recent advances in large language models (LLMs) show potential for clinical applications, such as clinical decision support and trial recommendations. However, the GPT-4 LLM predicts an excessive number of ICD codes for medical coding tasks, leading to high recall but low precision. To tackle this challenge, we introduce LLM-codex, a two-stage approach to predict ICD codes that first generates evidence proposals using an LLM and then employs an LSTM-based verification stage. The LSTM learns from both the LLM's high recall and human expert's high precision, using a custom loss function. Our model is the only approach that simultaneously achieves state-of-the-art results in medical coding accuracy, accuracy on rare codes, and sentence-level evidence identification to support coding decisions without training on human-annotated evidence according to experiments on the MIMIC dataset.
Extend and Explain: Interpreting Very Long Language Models
Sep 07, 2022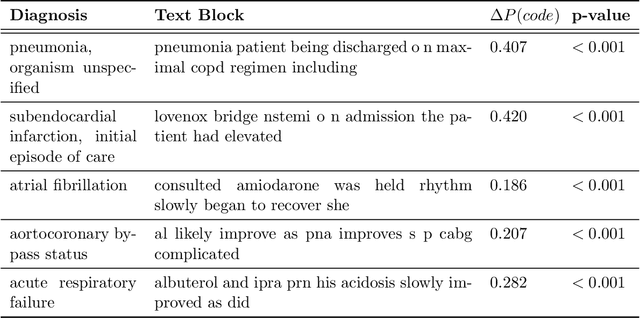
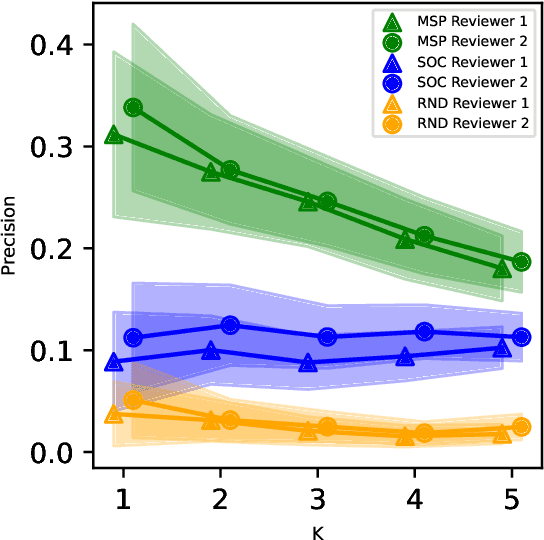
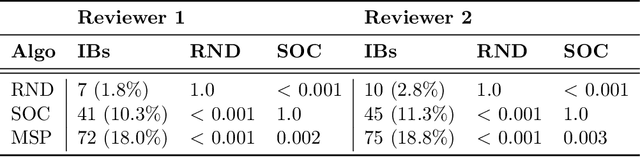
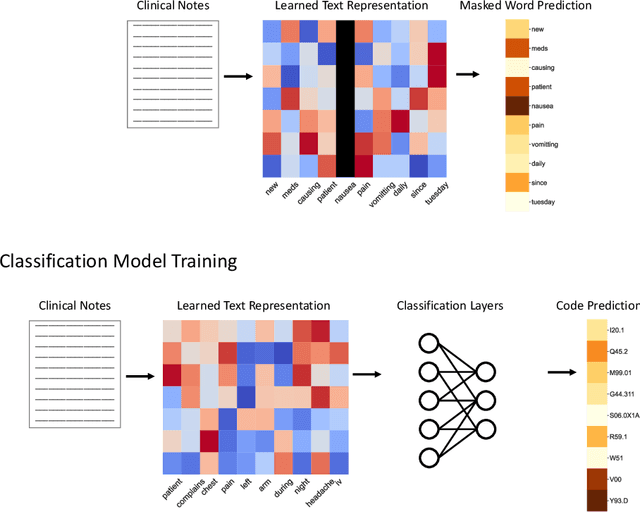
Abstract:While Transformer language models (LMs) are state-of-the-art for information extraction, long text introduces computational challenges requiring suboptimal preprocessing steps or alternative model architectures. Sparse-attention LMs can represent longer sequences, overcoming performance hurdles. However, it remains unclear how to explain predictions from these models, as not all tokens attend to each other in the self-attention layers, and long sequences pose computational challenges for explainability algorithms when runtime depends on document length. These challenges are severe in the medical context where documents can be very long, and machine learning (ML) models must be auditable and trustworthy. We introduce a novel Masked Sampling Procedure (MSP) to identify the text blocks that contribute to a prediction, apply MSP in the context of predicting diagnoses from medical text, and validate our approach with a blind review by two clinicians. Our method identifies about 1.7x more clinically informative text blocks than the previous state-of-the-art, runs up to 100x faster, and is tractable for generating important phrase pairs. MSP is particularly well-suited to long LMs but can be applied to any text classifier. We provide a general implementation of MSP.
ReFACTor: Practical Low-Rank Matrix Estimation Under Column-Sparsity
May 22, 2017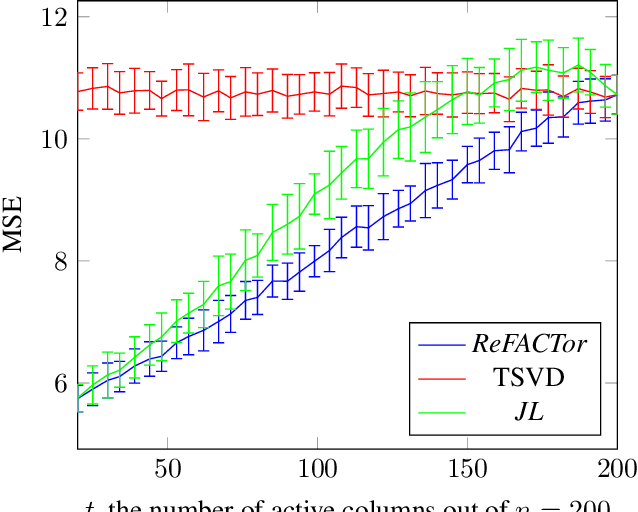
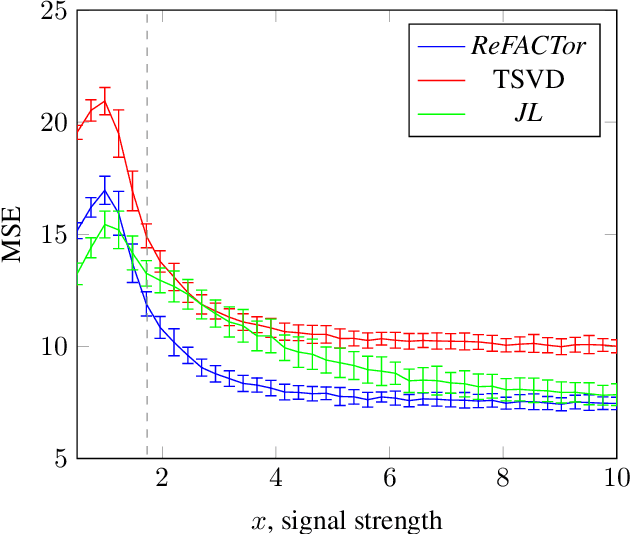
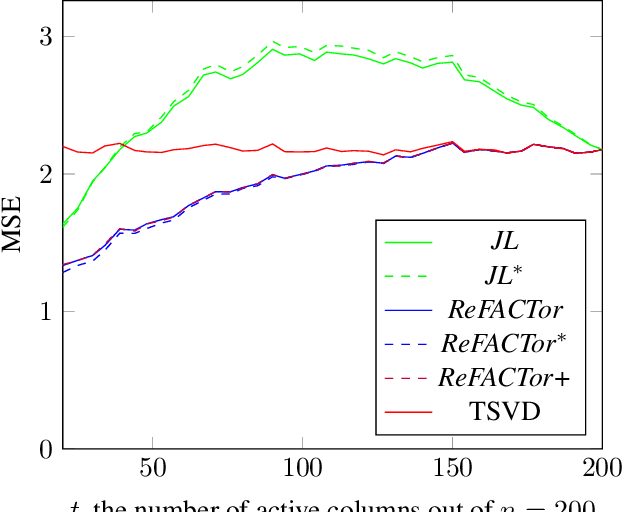
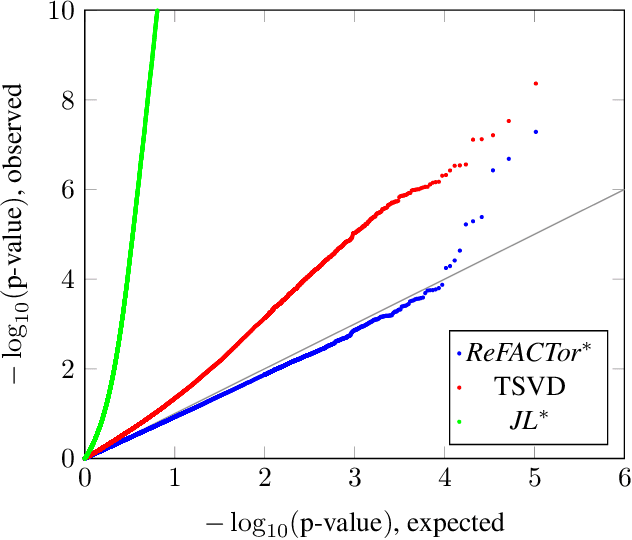
Abstract:Various problems in data analysis and statistical genetics call for recovery of a column-sparse, low-rank matrix from noisy observations. We propose ReFACTor, a simple variation of the classical Truncated Singular Value Decomposition (TSVD) algorithm. In contrast to previous sparse principal component analysis (PCA) algorithms, our algorithm can provably reveal a low-rank signal matrix better, and often significantly better, than the widely used TSVD, making it the algorithm of choice whenever column-sparsity is suspected. Empirically, we observe that ReFACTor consistently outperforms TSVD even when the underlying signal is not sparse, suggesting that it is generally safe to use ReFACTor instead of TSVD and PCA. The algorithm is extremely simple to implement and its running time is dominated by the runtime of PCA, making it as practical as standard principal component analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge