Jeffrey Hertzberg
XAIQA: Explainer-Based Data Augmentation for Extractive Question Answering
Dec 06, 2023Abstract:Extractive question answering (QA) systems can enable physicians and researchers to query medical records, a foundational capability for designing clinical studies and understanding patient medical history. However, building these systems typically requires expert-annotated QA pairs. Large language models (LLMs), which can perform extractive QA, depend on high quality data in their prompts, specialized for the application domain. We introduce a novel approach, XAIQA, for generating synthetic QA pairs at scale from data naturally available in electronic health records. Our method uses the idea of a classification model explainer to generate questions and answers about medical concepts corresponding to medical codes. In an expert evaluation with two physicians, our method identifies $2.2\times$ more semantic matches and $3.8\times$ more clinical abbreviations than two popular approaches that use sentence transformers to create QA pairs. In an ML evaluation, adding our QA pairs improves performance of GPT-4 as an extractive QA model, including on difficult questions. In both the expert and ML evaluations, we examine trade-offs between our method and sentence transformers for QA pair generation depending on question difficulty.
Extend and Explain: Interpreting Very Long Language Models
Sep 07, 2022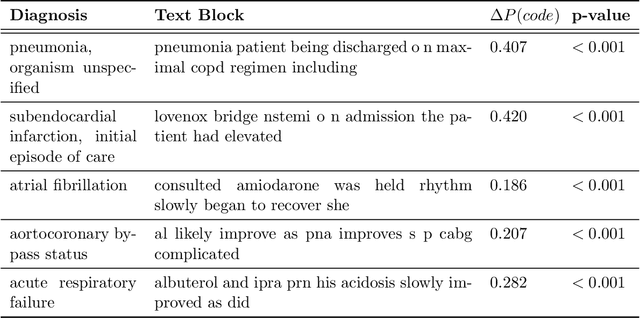
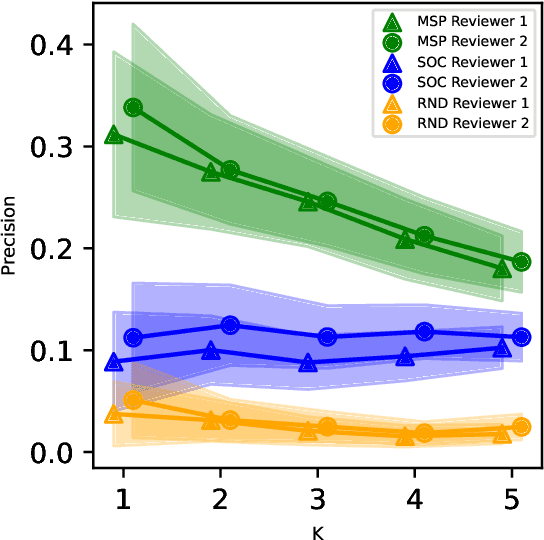
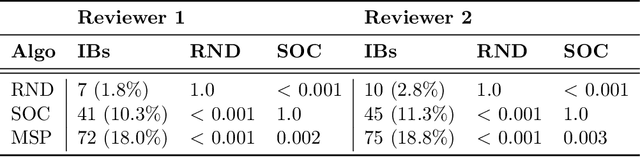
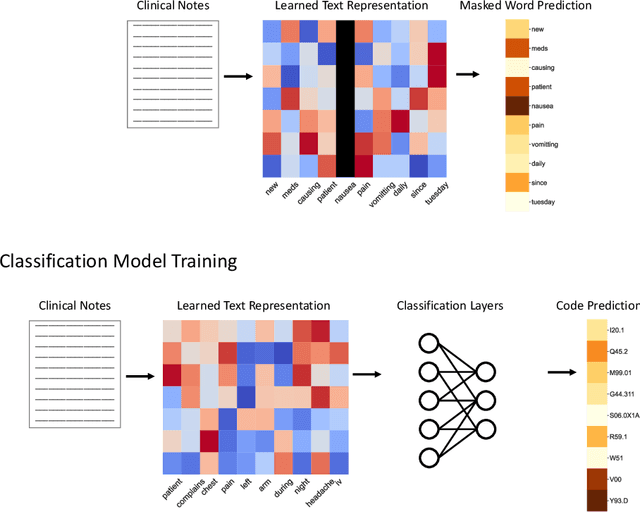
Abstract:While Transformer language models (LMs) are state-of-the-art for information extraction, long text introduces computational challenges requiring suboptimal preprocessing steps or alternative model architectures. Sparse-attention LMs can represent longer sequences, overcoming performance hurdles. However, it remains unclear how to explain predictions from these models, as not all tokens attend to each other in the self-attention layers, and long sequences pose computational challenges for explainability algorithms when runtime depends on document length. These challenges are severe in the medical context where documents can be very long, and machine learning (ML) models must be auditable and trustworthy. We introduce a novel Masked Sampling Procedure (MSP) to identify the text blocks that contribute to a prediction, apply MSP in the context of predicting diagnoses from medical text, and validate our approach with a blind review by two clinicians. Our method identifies about 1.7x more clinically informative text blocks than the previous state-of-the-art, runs up to 100x faster, and is tractable for generating important phrase pairs. MSP is particularly well-suited to long LMs but can be applied to any text classifier. We provide a general implementation of MSP.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge