Yijia Xiao
BioVerge: A Comprehensive Benchmark and Study of Self-Evaluating Agents for Biomedical Hypothesis Generation
Nov 12, 2025Abstract:Hypothesis generation in biomedical research has traditionally centered on uncovering hidden relationships within vast scientific literature, often using methods like Literature-Based Discovery (LBD). Despite progress, current approaches typically depend on single data types or predefined extraction patterns, which restricts the discovery of novel and complex connections. Recent advances in Large Language Model (LLM) agents show significant potential, with capabilities in information retrieval, reasoning, and generation. However, their application to biomedical hypothesis generation has been limited by the absence of standardized datasets and execution environments. To address this, we introduce BioVerge, a comprehensive benchmark, and BioVerge Agent, an LLM-based agent framework, to create a standardized environment for exploring biomedical hypothesis generation at the frontier of existing scientific knowledge. Our dataset includes structured and textual data derived from historical biomedical hypotheses and PubMed literature, organized to support exploration by LLM agents. BioVerge Agent utilizes a ReAct-based approach with distinct Generation and Evaluation modules that iteratively produce and self-assess hypothesis proposals. Through extensive experimentation, we uncover key insights: 1) different architectures of BioVerge Agent influence exploration diversity and reasoning strategies; 2) structured and textual information sources each provide unique, critical contexts that enhance hypothesis generation; and 3) self-evaluation significantly improves the novelty and relevance of proposed hypotheses.
Advancing AI-Scientist Understanding: Making LLM Think Like a Physicist with Interpretable Reasoning
Apr 02, 2025Abstract:Large Language Models (LLMs) are playing an expanding role in physics research by enhancing reasoning, symbolic manipulation, and numerical computation. However, ensuring the reliability and interpretability of their outputs remains a significant challenge. In our framework, we conceptualize the collaboration between AI and human scientists as a dynamic interplay among three modules: the reasoning module, the interpretation module, and the AI-scientist interaction module. Recognizing that effective physics reasoning demands rigorous logical consistency, quantitative precision, and deep integration with established theoretical models, we introduce the interpretation module to improve the understanding of AI-generated outputs, which is not previously explored in the literature. This module comprises multiple specialized agents, including summarizers, model builders, UI builders, and testers, which collaboratively structure LLM outputs within a physically grounded framework, by constructing a more interpretable science model. A case study demonstrates that our approach enhances transparency, facilitates validation, and strengthens AI-augmented reasoning in scientific discovery.
Protein Large Language Models: A Comprehensive Survey
Feb 21, 2025Abstract:Protein-specific large language models (Protein LLMs) are revolutionizing protein science by enabling more efficient protein structure prediction, function annotation, and design. While existing surveys focus on specific aspects or applications, this work provides the first comprehensive overview of Protein LLMs, covering their architectures, training datasets, evaluation metrics, and diverse applications. Through a systematic analysis of over 100 articles, we propose a structured taxonomy of state-of-the-art Protein LLMs, analyze how they leverage large-scale protein sequence data for improved accuracy, and explore their potential in advancing protein engineering and biomedical research. Additionally, we discuss key challenges and future directions, positioning Protein LLMs as essential tools for scientific discovery in protein science. Resources are maintained at https://github.com/Yijia-Xiao/Protein-LLM-Survey.
CSR-Bench: Benchmarking LLM Agents in Deployment of Computer Science Research Repositories
Feb 10, 2025



Abstract:The increasing complexity of computer science research projects demands more effective tools for deploying code repositories. Large Language Models (LLMs), such as Anthropic Claude and Meta Llama, have demonstrated significant advancements across various fields of computer science research, including the automation of diverse software engineering tasks. To evaluate the effectiveness of LLMs in handling complex code development tasks of research projects, particularly for NLP/CV/AI/ML/DM topics, we introduce CSR-Bench, a benchmark for Computer Science Research projects. This benchmark assesses LLMs from various aspects including accuracy, efficiency, and deployment script quality, aiming to explore their potential in conducting computer science research autonomously. We also introduce a novel framework, CSR-Agents, that utilizes multiple LLM agents to automate the deployment of GitHub code repositories of computer science research projects. Specifically, by checking instructions from markdown files and interpreting repository structures, the model generates and iteratively improves bash commands that set up the experimental environments and deploy the code to conduct research tasks. Preliminary results from CSR-Bench indicate that LLM agents can significantly enhance the workflow of repository deployment, thereby boosting developer productivity and improving the management of developmental workflows.
Memorize and Rank: Elevating Large Language Models for Clinical Diagnosis Prediction
Jan 28, 2025Abstract:Clinical diagnosis prediction models, when provided with a patient's medical history, aim to detect potential diseases early, facilitating timely intervention and improving prognostic outcomes. However, the inherent scarcity of patient data and large disease candidate space often pose challenges in developing satisfactory models for this intricate task. The exploration of leveraging Large Language Models (LLMs) for encapsulating clinical decision processes has been limited. We introduce MERA, a clinical diagnosis prediction model that bridges pertaining natural language knowledge with medical practice. We apply hierarchical contrastive learning on a disease candidate ranking list to alleviate the large decision space issue. With concept memorization through fine-tuning, we bridge the natural language clinical knowledge with medical codes. Experimental results on MIMIC-III and IV datasets show that MERA achieves the state-of-the-art diagnosis prediction performance and dramatically elevates the diagnosis prediction capabilities of generative LMs.
TradingAgents: Multi-Agents LLM Financial Trading Framework
Dec 28, 2024



Abstract:Significant progress has been made in automated problem-solving using societies of agents powered by large language models (LLMs). In finance, efforts have largely focused on single-agent systems handling specific tasks or multi-agent frameworks independently gathering data. However, multi-agent systems' potential to replicate real-world trading firms' collaborative dynamics remains underexplored. TradingAgents proposes a novel stock trading framework inspired by trading firms, featuring LLM-powered agents in specialized roles such as fundamental analysts, sentiment analysts, technical analysts, and traders with varied risk profiles. The framework includes Bull and Bear researcher agents assessing market conditions, a risk management team monitoring exposure, and traders synthesizing insights from debates and historical data to make informed decisions. By simulating a dynamic, collaborative trading environment, this framework aims to improve trading performance. Detailed architecture and extensive experiments reveal its superiority over baseline models, with notable improvements in cumulative returns, Sharpe ratio, and maximum drawdown, highlighting the potential of multi-agent LLM frameworks in financial trading.
Scito2M: A 2 Million, 30-Year Cross-disciplinary Dataset for Temporal Scientometric Analysis
Oct 12, 2024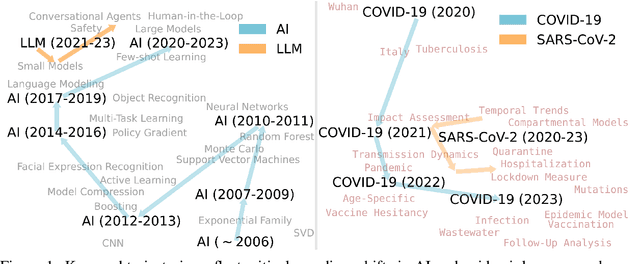
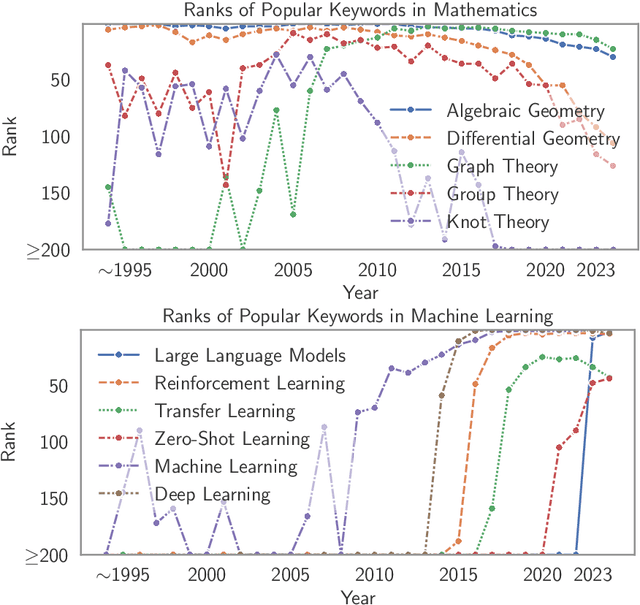

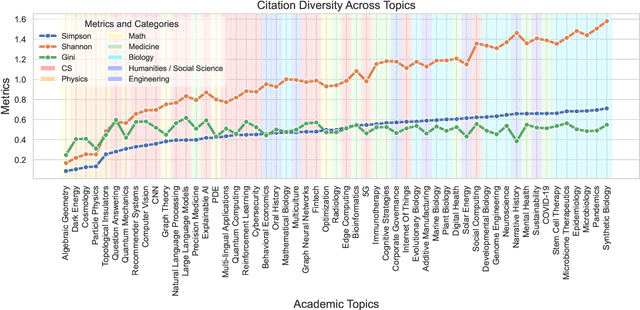
Abstract:Understanding the creation, evolution, and dissemination of scientific knowledge is crucial for bridging diverse subject areas and addressing complex global challenges such as pandemics, climate change, and ethical AI. Scientometrics, the quantitative and qualitative study of scientific literature, provides valuable insights into these processes. We introduce Scito2M, a longitudinal scientometric dataset with over two million academic publications, providing comprehensive contents information and citation graphs to support cross-disciplinary analyses. Using Scito2M, we conduct a temporal study spanning over 30 years to explore key questions in scientometrics: the evolution of academic terminology, citation patterns, and interdisciplinary knowledge exchange. Our findings reveal critical insights, such as disparities in epistemic cultures, knowledge production modes, and citation practices. For example, rapidly developing, application-driven fields like LLMs exhibit significantly shorter citation age (2.48 years) compared to traditional theoretical disciplines like oral history (9.71 years).
ProteinGPT: Multimodal LLM for Protein Property Prediction and Structure Understanding
Aug 21, 2024Abstract:Understanding biological processes, drug development, and biotechnological advancements requires detailed analysis of protein structures and sequences, a task in protein research that is inherently complex and time-consuming when performed manually. To streamline this process, we introduce ProteinGPT, a state-of-the-art multi-modal protein chat system, that allows users to upload protein sequences and/or structures for comprehensive protein analysis and responsive inquiries. ProteinGPT seamlessly integrates protein sequence and structure encoders with linear projection layers for precise representation adaptation, coupled with a large language model (LLM) to generate accurate and contextually relevant responses. To train ProteinGPT, we construct a large-scale dataset of 132,092 proteins with annotations, and optimize the instruction-tuning process using GPT-4o. This innovative system ensures accurate alignment between the user-uploaded data and prompts, simplifying protein analysis. Experiments show that ProteinGPT can produce promising responses to proteins and their corresponding questions.
LogicVista: Multimodal LLM Logical Reasoning Benchmark in Visual Contexts
Jul 06, 2024Abstract:We propose LogicVista, an evaluation benchmark that assesses the integrated logical reasoning capabilities of multimodal large language models (MLLMs) in Visual contexts. Recent advancements in MLLMs have demonstrated various fascinating abilities, from crafting poetry based on an image to performing mathematical reasoning. However, there is still a lack of systematic evaluation of MLLMs' proficiency in logical reasoning tasks, which are essential for activities like navigation and puzzle-solving. Thus we evaluate general logical cognition abilities across 5 logical reasoning tasks encompassing 9 different capabilities, using a sample of 448 multiple-choice questions. Each question is annotated with the correct answer and the human-written reasoning behind the selection, enabling both open-ended and multiple-choice evaluation. A total of 8 MLLMs are comprehensively evaluated using LogicVista. Code and Data Available at https://github.com/Yijia-Xiao/LogicVista.
Geneverse: A collection of Open-source Multimodal Large Language Models for Genomic and Proteomic Research
Jun 21, 2024Abstract:The applications of large language models (LLMs) are promising for biomedical and healthcare research. Despite the availability of open-source LLMs trained using a wide range of biomedical data, current research on the applications of LLMs to genomics and proteomics is still limited. To fill this gap, we propose a collection of finetuned LLMs and multimodal LLMs (MLLMs), known as Geneverse, for three novel tasks in genomic and proteomic research. The models in Geneverse are trained and evaluated based on domain-specific datasets, and we use advanced parameter-efficient finetuning techniques to achieve the model adaptation for tasks including the generation of descriptions for gene functions, protein function inference from its structure, and marker gene selection from spatial transcriptomic data. We demonstrate that adapted LLMs and MLLMs perform well for these tasks and may outperform closed-source large-scale models based on our evaluations focusing on both truthfulness and structural correctness. All of the training strategies and base models we used are freely accessible.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge