Quanquan Gu
Protein Autoregressive Modeling via Multiscale Structure Generation
Feb 04, 2026Abstract:We present protein autoregressive modeling (PAR), the first multi-scale autoregressive framework for protein backbone generation via coarse-to-fine next-scale prediction. Using the hierarchical nature of proteins, PAR generates structures that mimic sculpting a statue, forming a coarse topology and refining structural details over scales. To achieve this, PAR consists of three key components: (i) multi-scale downsampling operations that represent protein structures across multiple scales during training; (ii) an autoregressive transformer that encodes multi-scale information and produces conditional embeddings to guide structure generation; (iii) a flow-based backbone decoder that generates backbone atoms conditioned on these embeddings. Moreover, autoregressive models suffer from exposure bias, caused by the training and the generation procedure mismatch, and substantially degrades structure generation quality. We effectively alleviate this issue by adopting noisy context learning and scheduled sampling, enabling robust backbone generation. Notably, PAR exhibits strong zero-shot generalization, supporting flexible human-prompted conditional generation and motif scaffolding without requiring fine-tuning. On the unconditional generation benchmark, PAR effectively learns protein distributions and produces backbones of high design quality, and exhibits favorable scaling behavior. Together, these properties establish PAR as a promising framework for protein structure generation.
Scalable Spatio-Temporal SE(3) Diffusion for Long-Horizon Protein Dynamics
Feb 02, 2026Abstract:Molecular dynamics (MD) simulations remain the gold standard for studying protein dynamics, but their computational cost limits access to biologically relevant timescales. Recent generative models have shown promise in accelerating simulations, yet they struggle with long-horizon generation due to architectural constraints, error accumulation, and inadequate modeling of spatio-temporal dynamics. We present STAR-MD (Spatio-Temporal Autoregressive Rollout for Molecular Dynamics), a scalable SE(3)-equivariant diffusion model that generates physically plausible protein trajectories over microsecond timescales. Our key innovation is a causal diffusion transformer with joint spatio-temporal attention that efficiently captures complex space-time dependencies while avoiding the memory bottlenecks of existing methods. On the standard ATLAS benchmark, STAR-MD achieves state-of-the-art performance across all metrics--substantially improving conformational coverage, structural validity, and dynamic fidelity compared to previous methods. STAR-MD successfully extrapolates to generate stable microsecond-scale trajectories where baseline methods fail catastrophically, maintaining high structural quality throughout the extended rollout. Our comprehensive evaluation reveals severe limitations in current models for long-horizon generation, while demonstrating that STAR-MD's joint spatio-temporal modeling enables robust dynamics simulation at biologically relevant timescales, paving the way for accelerated exploration of protein function.
Deep Delta Learning
Jan 01, 2026Abstract:The efficacy of deep residual networks is fundamentally predicated on the identity shortcut connection. While this mechanism effectively mitigates the vanishing gradient problem, it imposes a strictly additive inductive bias on feature transformations, thereby limiting the network's capacity to model complex state transitions. In this paper, we introduce Deep Delta Learning (DDL), a novel architecture that generalizes the standard residual connection by modulating the identity shortcut with a learnable, data-dependent geometric transformation. This transformation, termed the Delta Operator, constitutes a rank-1 perturbation of the identity matrix, parameterized by a reflection direction vector $\mathbf{k}(\mathbf{X})$ and a gating scalar $β(\mathbf{X})$. We provide a spectral analysis of this operator, demonstrating that the gate $β(\mathbf{X})$ enables dynamic interpolation between identity mapping, orthogonal projection, and geometric reflection. Furthermore, we restructure the residual update as a synchronous rank-1 injection, where the gate acts as a dynamic step size governing both the erasure of old information and the writing of new features. This unification empowers the network to explicitly control the spectrum of its layer-wise transition operator, enabling the modeling of complex, non-monotonic dynamics while preserving the stable training characteristics of gated residual architectures.
Group Representational Position Encoding
Dec 08, 2025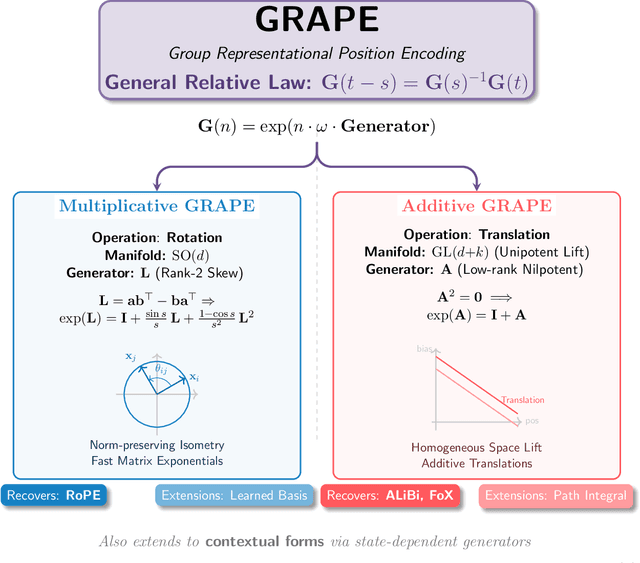
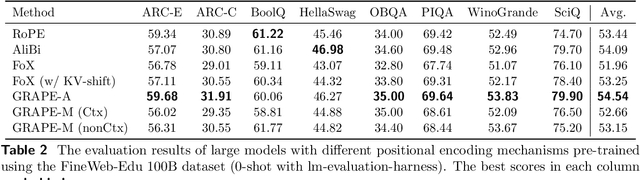
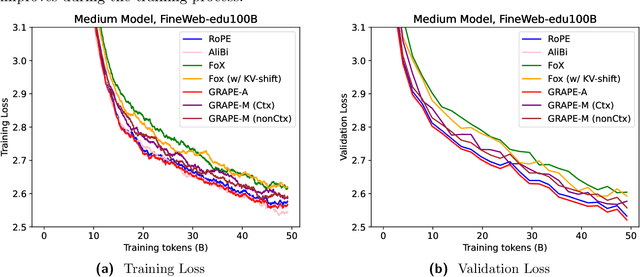
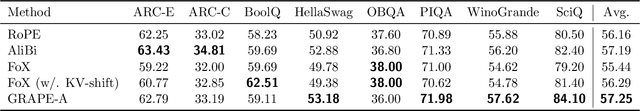
Abstract:We present GRAPE (Group RepresentAtional Position Encoding), a unified framework for positional encoding based on group actions. GRAPE brings together two families of mechanisms: (i) multiplicative rotations (Multiplicative GRAPE) in $\mathrm{SO}(d)$ and (ii) additive logit biases (Additive GRAPE) arising from unipotent actions in the general linear group $\mathrm{GL}$. In Multiplicative GRAPE, a position $n \in \mathbb{Z}$ (or $t \in \mathbb{R}$) acts as $\mathbf{G}(n)=\exp(n\,ω\,\mathbf{L})$ with a rank-2 skew generator $\mathbf{L} \in \mathbb{R}^{d \times d}$, yielding a relative, compositional, norm-preserving map with a closed-form matrix exponential. RoPE is recovered exactly when the $d/2$ planes are the canonical coordinate pairs with log-uniform spectrum. Learned commuting subspaces and compact non-commuting mixtures strictly extend this geometry to capture cross-subspace feature coupling at $O(d)$ and $O(r d)$ cost per head, respectively. In Additive GRAPE, additive logits arise as rank-1 (or low-rank) unipotent actions, recovering ALiBi and the Forgetting Transformer (FoX) as exact special cases while preserving an exact relative law and streaming cacheability. Altogether, GRAPE supplies a principled design space for positional geometry in long-context models, subsuming RoPE and ALiBi as special cases. Project Page: https://github.com/model-architectures/GRAPE.
Higher-order Linear Attention
Oct 31, 2025Abstract:The quadratic cost of scaled dot-product attention is a central obstacle to scaling autoregressive language models to long contexts. Linear-time attention and State Space Models (SSMs) provide scalable alternatives but are typically restricted to first-order or kernel-based approximations, which can limit expressivity. We introduce Higher-order Linear Attention (HLA), a causal, streaming mechanism that realizes higher interactions via compact prefix sufficient statistics. In the second-order case, HLA maintains a constant-size state and computes per-token outputs in linear time without materializing any $n \times n$ matrices. We give closed-form streaming identities, a strictly causal masked variant using two additional summaries, and a chunk-parallel training scheme based on associative scans that reproduces the activations of a serial recurrence exactly. We further outline extensions to third and higher orders. Collectively, these results position HLA as a principled, scalable building block that combines attention-like, data-dependent mixing with the efficiency of modern recurrent architectures. Project Page: https://github.com/yifanzhang-pro/HLA.
Causal Attention with Lookahead Keys
Sep 09, 2025Abstract:In standard causal attention, each token's query, key, and value (QKV) are static and encode only preceding context. We introduce CAuSal aTtention with Lookahead kEys (CASTLE), an attention mechanism that continually updates each token's keys as the context unfolds. We term these updated keys lookahead keys because they belong to earlier positions yet integrate information from tokens that appear later relative to those positions, while strictly preserving the autoregressive property. Although the mechanism appears sequential, we derive a mathematical equivalence that avoids explicitly materializing lookahead keys at each position and enables efficient parallel training. On language modeling benchmarks, CASTLE consistently outperforms standard causal attention across model scales, reducing validation perplexity and improving performance on a range of downstream tasks.
SwingArena: Competitive Programming Arena for Long-context GitHub Issue Solving
May 29, 2025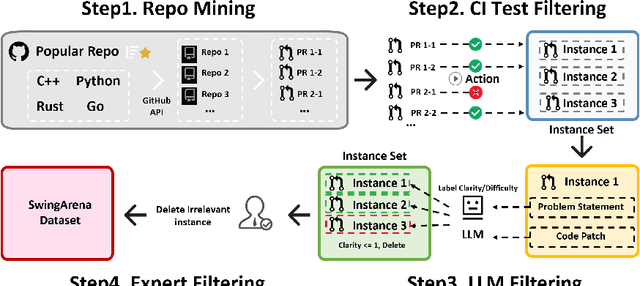
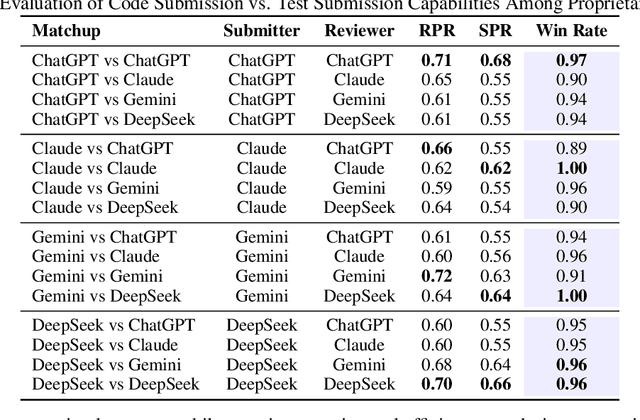
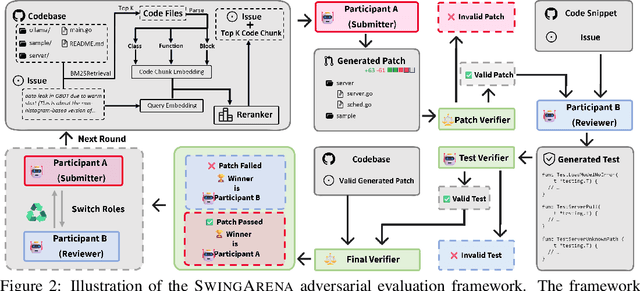
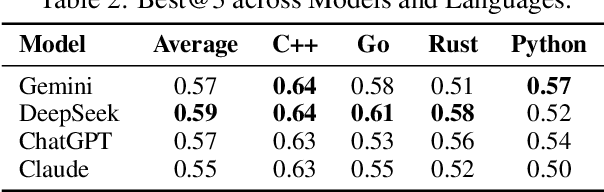
Abstract:We present SwingArena, a competitive evaluation framework for Large Language Models (LLMs) that closely mirrors real-world software development workflows. Unlike traditional static benchmarks, SwingArena models the collaborative process of software iteration by pairing LLMs as submitters, who generate patches, and reviewers, who create test cases and verify the patches through continuous integration (CI) pipelines. To support these interactive evaluations, we introduce a retrieval-augmented code generation (RACG) module that efficiently handles long-context challenges by providing syntactically and semantically relevant code snippets from large codebases, supporting multiple programming languages (C++, Python, Rust, and Go). This enables the framework to scale across diverse tasks and contexts while respecting token limitations. Our experiments, using over 400 high-quality real-world GitHub issues selected from a pool of 2,300 issues, show that models like GPT-4o excel at aggressive patch generation, whereas DeepSeek and Gemini prioritize correctness in CI validation. SwingArena presents a scalable and extensible methodology for evaluating LLMs in realistic, CI-driven software development settings. More details are available on our project page: swing-bench.github.io
Designing Cyclic Peptides via Harmonic SDE with Atom-Bond Modeling
May 27, 2025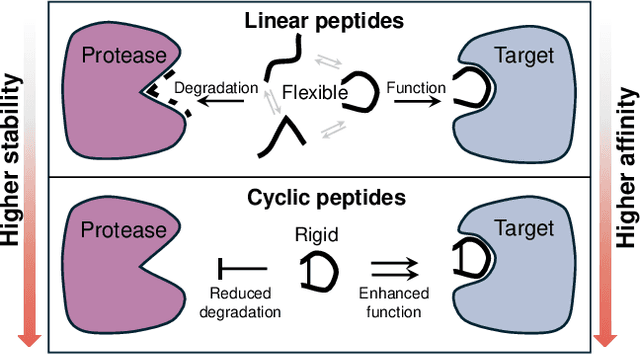
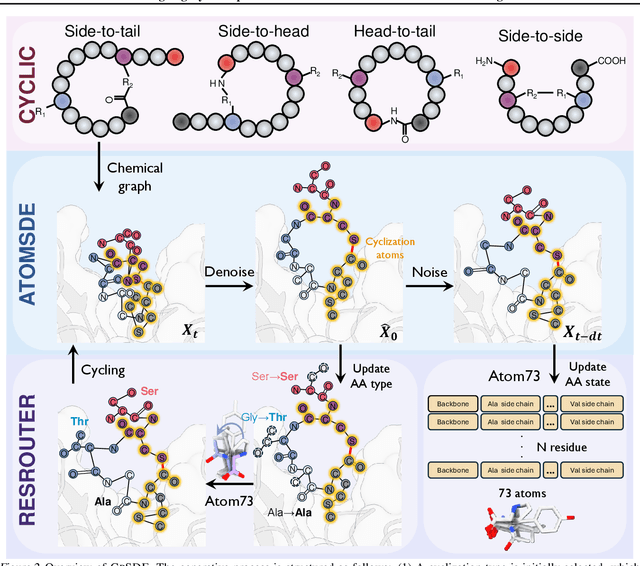
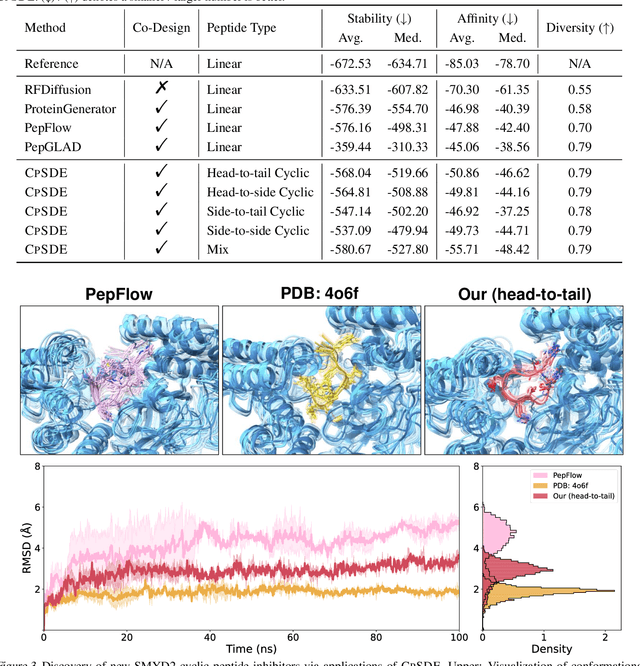
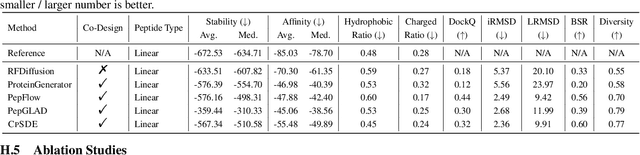
Abstract:Cyclic peptides offer inherent advantages in pharmaceuticals. For example, cyclic peptides are more resistant to enzymatic hydrolysis compared to linear peptides and usually exhibit excellent stability and affinity. Although deep generative models have achieved great success in linear peptide design, several challenges prevent the development of computational methods for designing diverse types of cyclic peptides. These challenges include the scarcity of 3D structural data on target proteins and associated cyclic peptide ligands, the geometric constraints that cyclization imposes, and the involvement of non-canonical amino acids in cyclization. To address the above challenges, we introduce CpSDE, which consists of two key components: AtomSDE, a generative structure prediction model based on harmonic SDE, and ResRouter, a residue type predictor. Utilizing a routed sampling algorithm that alternates between these two models to iteratively update sequences and structures, CpSDE facilitates the generation of cyclic peptides. By employing explicit all-atom and bond modeling, CpSDE overcomes existing data limitations and is proficient in designing a wide variety of cyclic peptides. Our experimental results demonstrate that the cyclic peptides designed by our method exhibit reliable stability and affinity.
Simultaneous Modeling of Protein Conformation and Dynamics via Autoregression
May 23, 2025Abstract:Understanding protein dynamics is critical for elucidating their biological functions. The increasing availability of molecular dynamics (MD) data enables the training of deep generative models to efficiently explore the conformational space of proteins. However, existing approaches either fail to explicitly capture the temporal dependencies between conformations or do not support direct generation of time-independent samples. To address these limitations, we introduce ConfRover, an autoregressive model that simultaneously learns protein conformation and dynamics from MD trajectories, supporting both time-dependent and time-independent sampling. At the core of our model is a modular architecture comprising: (i) an encoding layer, adapted from protein folding models, that embeds protein-specific information and conformation at each time frame into a latent space; (ii) a temporal module, a sequence model that captures conformational dynamics across frames; and (iii) an SE(3) diffusion model as the structure decoder, generating conformations in continuous space. Experiments on ATLAS, a large-scale protein MD dataset of diverse structures, demonstrate the effectiveness of our model in learning conformational dynamics and supporting a wide range of downstream tasks. ConfRover is the first model to sample both protein conformations and trajectories within a single framework, offering a novel and flexible approach for learning from protein MD data.
On the Design of KL-Regularized Policy Gradient Algorithms for LLM Reasoning
May 23, 2025Abstract:Policy gradient algorithms have been successfully applied to enhance the reasoning capabilities of large language models (LLMs). Despite the widespread use of Kullback-Leibler (KL) regularization in policy gradient algorithms to stabilize training, the systematic exploration of how different KL divergence formulations can be estimated and integrated into surrogate loss functions for online reinforcement learning (RL) presents a nuanced and systematically explorable design space. In this paper, we propose regularized policy gradient (RPG), a systematic framework for deriving and analyzing KL-regularized policy gradient methods in the online RL setting. We derive policy gradients and corresponding surrogate loss functions for objectives regularized by both forward and reverse KL divergences, considering both normalized and unnormalized policy distributions. Furthermore, we present derivations for fully differentiable loss functions as well as REINFORCE-style gradient estimators, accommodating diverse algorithmic needs. We conduct extensive experiments on RL for LLM reasoning using these methods, showing improved or competitive results in terms of training stability and performance compared to strong baselines such as GRPO, REINFORCE++, and DAPO. The code is available at https://github.com/complex-reasoning/RPG.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge