Hui Shen
OVD: On-policy Verbal Distillation
Jan 29, 2026Abstract:Knowledge distillation offers a promising path to transfer reasoning capabilities from large teacher models to efficient student models; however, existing token-level on-policy distillation methods require token-level alignment between the student and teacher models, which restricts the student model's exploration ability, prevent effective use of interactive environment feedback, and suffer from severe memory bottlenecks in reinforcement learning. We introduce On-policy Verbal Distillation (OVD), a memory-efficient framework that replaces token-level probability matching with trajectory matching using discrete verbal scores (0--9) from teacher models. OVD dramatically reduces memory consumption while enabling on-policy distillation from teacher models with verbal feedback, and avoids token-level alignment, allowing the student model to freely explore the output space. Extensive experiments on Web question answering and mathematical reasoning tasks show that OVD substantially outperforms existing methods, delivering up to +12.9% absolute improvement in average EM on Web Q&A tasks and a up to +25.7% gain on math benchmarks (when trained with only one random samples), while also exhibiting superior training efficiency. Our project page is available at https://OVD.github.io
Locate, Steer, and Improve: A Practical Survey of Actionable Mechanistic Interpretability in Large Language Models
Jan 20, 2026Abstract:Mechanistic Interpretability (MI) has emerged as a vital approach to demystify the opaque decision-making of Large Language Models (LLMs). However, existing reviews primarily treat MI as an observational science, summarizing analytical insights while lacking a systematic framework for actionable intervention. To bridge this gap, we present a practical survey structured around the pipeline: "Locate, Steer, and Improve." We formally categorize Localizing (diagnosis) and Steering (intervention) methods based on specific Interpretable Objects to establish a rigorous intervention protocol. Furthermore, we demonstrate how this framework enables tangible improvements in Alignment, Capability, and Efficiency, effectively operationalizing MI as an actionable methodology for model optimization. The curated paper list of this work is available at https://github.com/rattlesnakey/Awesome-Actionable-MI-Survey.
MMFormalizer: Multimodal Autoformalization in the Wild
Jan 06, 2026Abstract:Autoformalization, which translates natural language mathematics into formal statements to enable machine reasoning, faces fundamental challenges in the wild due to the multimodal nature of the physical world, where physics requires inferring hidden constraints (e.g., mass or energy) from visual elements. To address this, we propose MMFormalizer, which extends autoformalization beyond text by integrating adaptive grounding with entities from real-world mathematical and physical domains. MMFormalizer recursively constructs formal propositions from perceptually grounded primitives through recursive grounding and axiom composition, with adaptive recursive termination ensuring that every abstraction is supported by visual evidence and anchored in dimensional or axiomatic grounding. We evaluate MMFormalizer on a new benchmark, PhyX-AF, comprising 115 curated samples from MathVerse, PhyX, Synthetic Geometry, and Analytic Geometry, covering diverse multimodal autoformalization tasks. Results show that frontier models such as GPT-5 and Gemini-3-Pro achieve the highest compile and semantic accuracy, with GPT-5 excelling in physical reasoning, while geometry remains the most challenging domain. Overall, MMFormalizer provides a scalable framework for unified multimodal autoformalization, bridging perception and formal reasoning. To the best of our knowledge, this is the first multimodal autoformalization method capable of handling classical mechanics (derived from the Hamiltonian), as well as relativity, quantum mechanics, and thermodynamics. More details are available on our project page: MMFormalizer.github.io
DoPE: Denoising Rotary Position Embedding
Nov 12, 2025Abstract:Rotary Position Embedding (RoPE) in Transformer models has inherent limits that weaken length extrapolation. We reinterpret the attention map with positional encoding as a noisy feature map, and propose Denoising Positional Encoding (DoPE), a training-free method based on truncated matrix entropy to detect outlier frequency bands in the feature map. Leveraging the noise characteristics of the feature map, we further reparameterize it with a parameter-free Gaussian distribution to achieve robust extrapolation. Our method theoretically reveals the underlying cause of the attention sink phenomenon and its connection to truncated matrix entropy. Experiments on needle-in-a-haystack and many-shot in-context learning tasks demonstrate that DoPE significantly improves retrieval accuracy and reasoning stability across extended contexts (up to 64K tokens). The results show that the denoising strategy for positional embeddings effectively mitigates attention sinks and restores balanced attention patterns, providing a simple yet powerful solution for improving length generalization. Our project page is Project: https://The-physical-picture-of-LLMs.github.io
PF-DAformer: Proximal Femur Segmentation via Domain Adaptive Transformer for Dual-Center QCT
Oct 30, 2025Abstract:Quantitative computed tomography (QCT) plays a crucial role in assessing bone strength and fracture risk by enabling volumetric analysis of bone density distribution in the proximal femur. However, deploying automated segmentation models in practice remains difficult because deep networks trained on one dataset often fail when applied to another. This failure stems from domain shift, where scanners, reconstruction settings, and patient demographics vary across institutions, leading to unstable predictions and unreliable quantitative metrics. Overcoming this barrier is essential for multi-center osteoporosis research and for ensuring that radiomics and structural finite element analysis results remain reproducible across sites. In this work, we developed a domain-adaptive transformer segmentation framework tailored for multi-institutional QCT. Our model is trained and validated on one of the largest hip fracture related research cohorts to date, comprising 1,024 QCT images scans from Tulane University and 384 scans from Rochester, Minnesota for proximal femur segmentation. To address domain shift, we integrate two complementary strategies within a 3D TransUNet backbone: adversarial alignment via Gradient Reversal Layer (GRL), which discourages the network from encoding site-specific cues, and statistical alignment via Maximum Mean Discrepancy (MMD), which explicitly reduces distributional mismatches between institutions. This dual mechanism balances invariance and fine-grained alignment, enabling scanner-agnostic feature learning while preserving anatomical detail.
A1: Asynchronous Test-Time Scaling via Conformal Prediction
Sep 18, 2025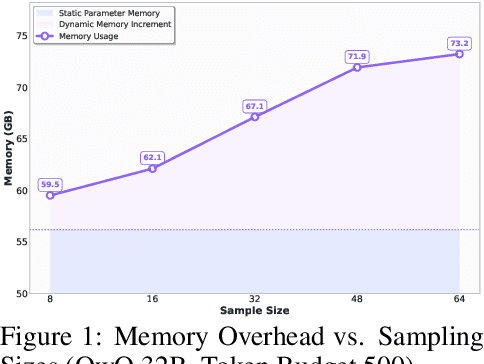
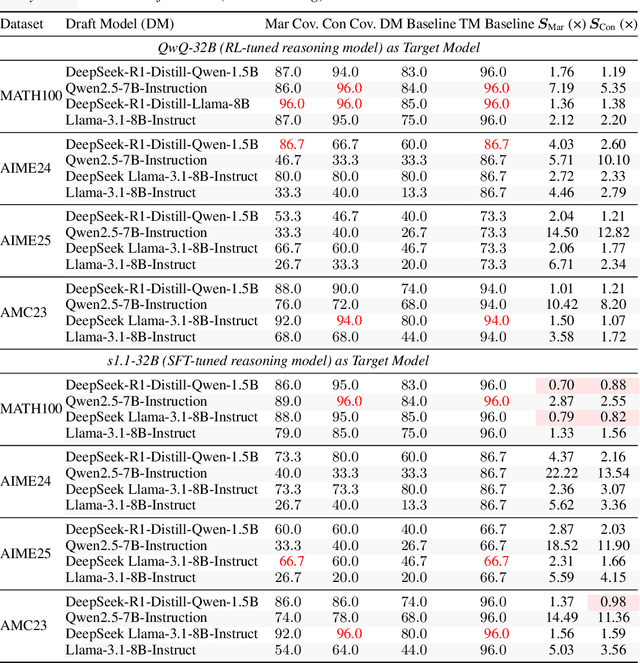
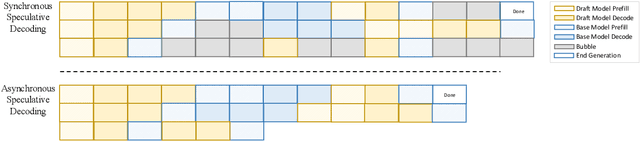
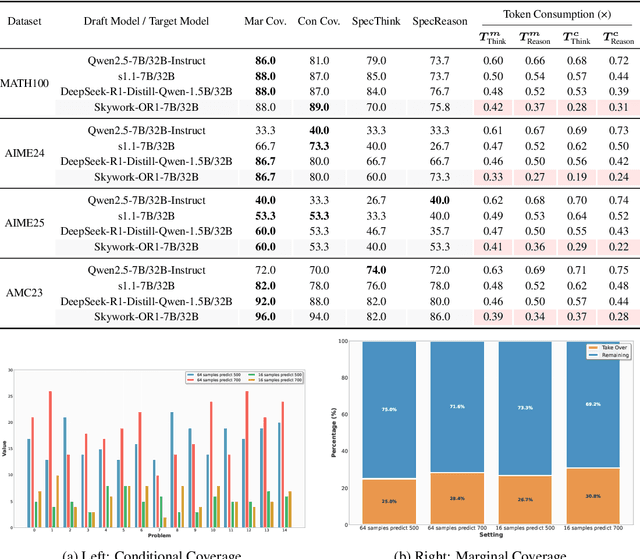
Abstract:Large language models (LLMs) benefit from test-time scaling, but existing methods face significant challenges, including severe synchronization overhead, memory bottlenecks, and latency, especially during speculative decoding with long reasoning chains. We introduce A1 (Asynchronous Test-Time Scaling), a statistically guaranteed adaptive inference framework that addresses these challenges. A1 refines arithmetic intensity to identify synchronization as the dominant bottleneck, proposes an online calibration strategy to enable asynchronous inference, and designs a three-stage rejection sampling pipeline that supports both sequential and parallel scaling. Through experiments on the MATH, AMC23, AIME24, and AIME25 datasets, across various draft-target model families, we demonstrate that A1 achieves a remarkable 56.7x speedup in test-time scaling and a 4.14x improvement in throughput, all while maintaining accurate rejection-rate control, reducing latency and memory overhead, and no accuracy loss compared to using target model scaling alone. These results position A1 as an efficient and principled solution for scalable LLM inference. We have released the code at https://github.com/menik1126/asynchronous-test-time-scaling.
LongEmotion: Measuring Emotional Intelligence of Large Language Models in Long-Context Interaction
Sep 09, 2025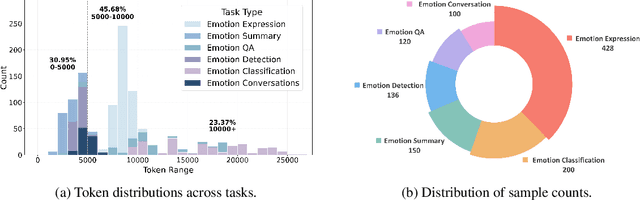
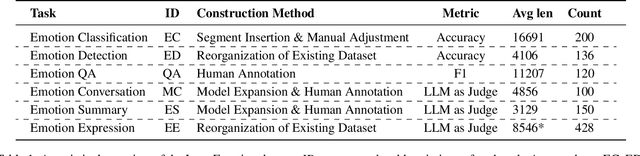
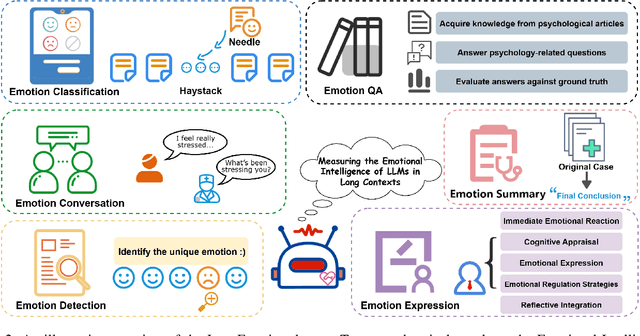
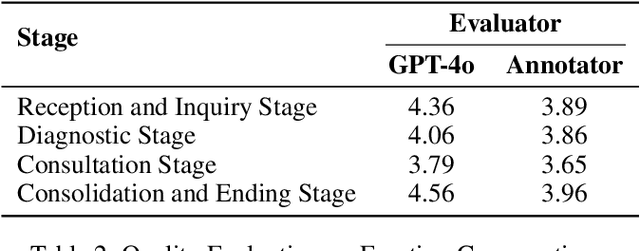
Abstract:Large language models (LLMs) make significant progress in Emotional Intelligence (EI) and long-context understanding. However, existing benchmarks tend to overlook certain aspects of EI in long-context scenarios, especially under realistic, practical settings where interactions are lengthy, diverse, and often noisy. To move towards such realistic settings, we present LongEmotion, a benchmark specifically designed for long-context EI tasks. It covers a diverse set of tasks, including Emotion Classification, Emotion Detection, Emotion QA, Emotion Conversation, Emotion Summary, and Emotion Expression. On average, the input length for these tasks reaches 8,777 tokens, with long-form generation required for Emotion Expression. To enhance performance under realistic constraints, we incorporate Retrieval-Augmented Generation (RAG) and Collaborative Emotional Modeling (CoEM), and compare them with standard prompt-based methods. Unlike conventional approaches, our RAG method leverages both the conversation context and the large language model itself as retrieval sources, avoiding reliance on external knowledge bases. The CoEM method further improves performance by decomposing the task into five stages, integrating both retrieval augmentation and limited knowledge injection. Experimental results show that both RAG and CoEM consistently enhance EI-related performance across most long-context tasks, advancing LLMs toward more practical and real-world EI applications. Furthermore, we conducted a comparative case study experiment on the GPT series to demonstrate the differences among various models in terms of EI. Code is available on GitHub at https://github.com/LongEmotion/LongEmotion, and the project page can be found at https://longemotion.github.io/.
AnchorDP3: 3D Affordance Guided Sparse Diffusion Policy for Robotic Manipulation
Jun 24, 2025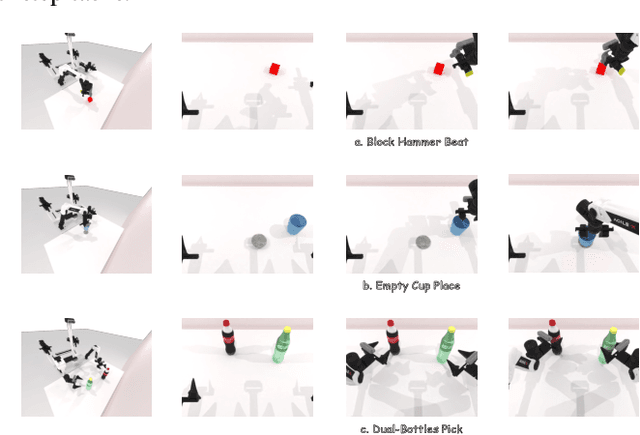
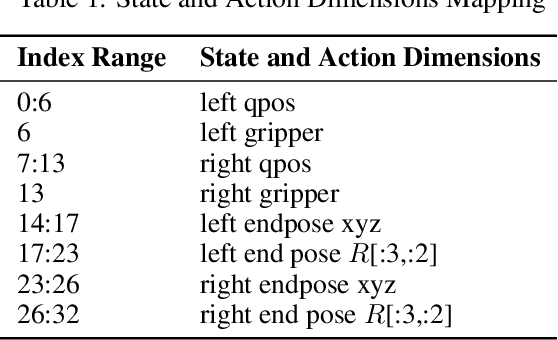
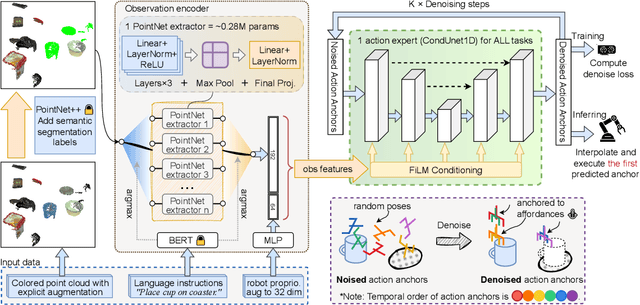
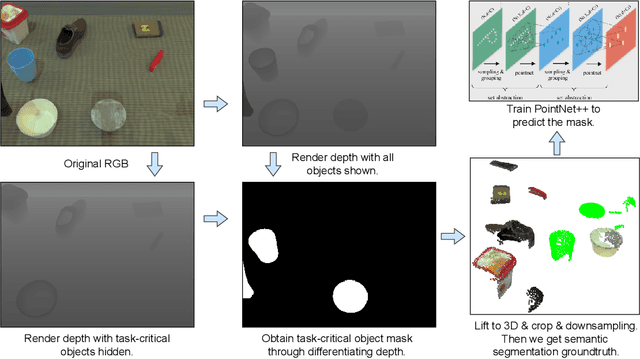
Abstract:We present AnchorDP3, a diffusion policy framework for dual-arm robotic manipulation that achieves state-of-the-art performance in highly randomized environments. AnchorDP3 integrates three key innovations: (1) Simulator-Supervised Semantic Segmentation, using rendered ground truth to explicitly segment task-critical objects within the point cloud, which provides strong affordance priors; (2) Task-Conditioned Feature Encoders, lightweight modules processing augmented point clouds per task, enabling efficient multi-task learning through a shared diffusion-based action expert; (3) Affordance-Anchored Keypose Diffusion with Full State Supervision, replacing dense trajectory prediction with sparse, geometrically meaningful action anchors, i.e., keyposes such as pre-grasp pose, grasp pose directly anchored to affordances, drastically simplifying the prediction space; the action expert is forced to predict both robot joint angles and end-effector poses simultaneously, which exploits geometric consistency to accelerate convergence and boost accuracy. Trained on large-scale, procedurally generated simulation data, AnchorDP3 achieves a 98.7% average success rate in the RoboTwin benchmark across diverse tasks under extreme randomization of objects, clutter, table height, lighting, and backgrounds. This framework, when integrated with the RoboTwin real-to-sim pipeline, has the potential to enable fully autonomous generation of deployable visuomotor policies from only scene and instruction, totally eliminating human demonstrations from learning manipulation skills.
SwingArena: Competitive Programming Arena for Long-context GitHub Issue Solving
May 29, 2025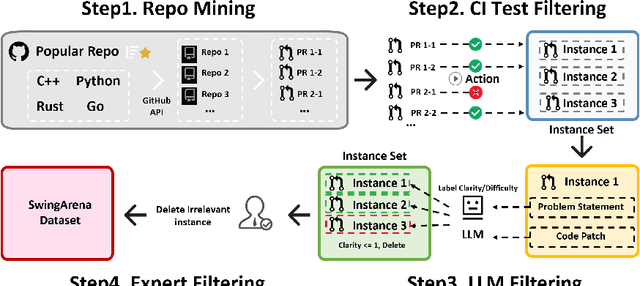
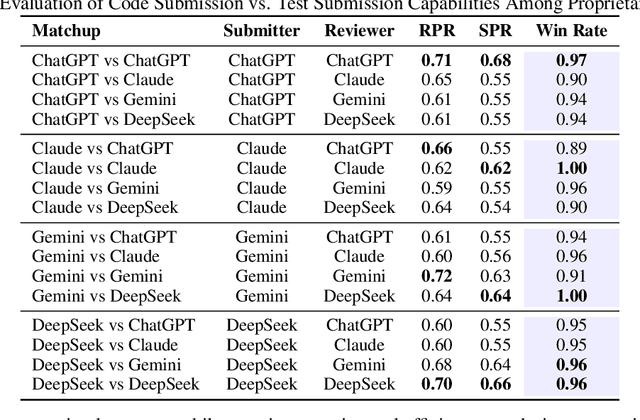
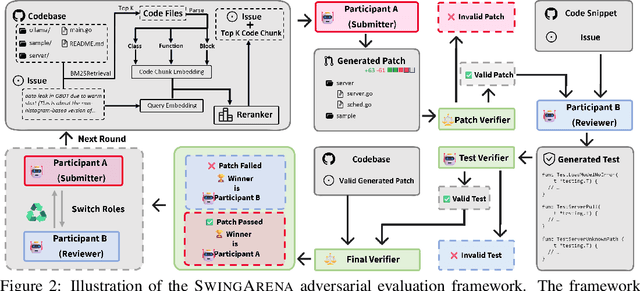
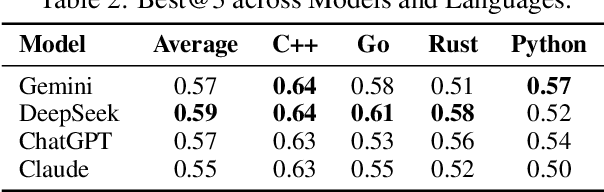
Abstract:We present SwingArena, a competitive evaluation framework for Large Language Models (LLMs) that closely mirrors real-world software development workflows. Unlike traditional static benchmarks, SwingArena models the collaborative process of software iteration by pairing LLMs as submitters, who generate patches, and reviewers, who create test cases and verify the patches through continuous integration (CI) pipelines. To support these interactive evaluations, we introduce a retrieval-augmented code generation (RACG) module that efficiently handles long-context challenges by providing syntactically and semantically relevant code snippets from large codebases, supporting multiple programming languages (C++, Python, Rust, and Go). This enables the framework to scale across diverse tasks and contexts while respecting token limitations. Our experiments, using over 400 high-quality real-world GitHub issues selected from a pool of 2,300 issues, show that models like GPT-4o excel at aggressive patch generation, whereas DeepSeek and Gemini prioritize correctness in CI validation. SwingArena presents a scalable and extensible methodology for evaluating LLMs in realistic, CI-driven software development settings. More details are available on our project page: swing-bench.github.io
Fully Spiking Neural Networks for Unified Frame-Event Object Tracking
May 27, 2025Abstract:The integration of image and event streams offers a promising approach for achieving robust visual object tracking in complex environments. However, current fusion methods achieve high performance at the cost of significant computational overhead and struggle to efficiently extract the sparse, asynchronous information from event streams, failing to leverage the energy-efficient advantages of event-driven spiking paradigms. To address this challenge, we propose the first fully Spiking Frame-Event Tracking framework called SpikeFET. This network achieves synergistic integration of convolutional local feature extraction and Transformer-based global modeling within the spiking paradigm, effectively fusing frame and event data. To overcome the degradation of translation invariance caused by convolutional padding, we introduce a Random Patchwork Module (RPM) that eliminates positional bias through randomized spatial reorganization and learnable type encoding while preserving residual structures. Furthermore, we propose a Spatial-Temporal Regularization (STR) strategy that overcomes similarity metric degradation from asymmetric features by enforcing spatio-temporal consistency among temporal template features in latent space. Extensive experiments across multiple benchmarks demonstrate that the proposed framework achieves superior tracking accuracy over existing methods while significantly reducing power consumption, attaining an optimal balance between performance and efficiency. The code will be released.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge