Guang Shi
Seedance 1.5 pro: A Native Audio-Visual Joint Generation Foundation Model
Dec 23, 2025Abstract:Recent strides in video generation have paved the way for unified audio-visual generation. In this work, we present Seedance 1.5 pro, a foundational model engineered specifically for native, joint audio-video generation. Leveraging a dual-branch Diffusion Transformer architecture, the model integrates a cross-modal joint module with a specialized multi-stage data pipeline, achieving exceptional audio-visual synchronization and superior generation quality. To ensure practical utility, we implement meticulous post-training optimizations, including Supervised Fine-Tuning (SFT) on high-quality datasets and Reinforcement Learning from Human Feedback (RLHF) with multi-dimensional reward models. Furthermore, we introduce an acceleration framework that boosts inference speed by over 10X. Seedance 1.5 pro distinguishes itself through precise multilingual and dialect lip-syncing, dynamic cinematic camera control, and enhanced narrative coherence, positioning it as a robust engine for professional-grade content creation. Seedance 1.5 pro is now accessible on Volcano Engine at https://console.volcengine.com/ark/region:ark+cn-beijing/experience/vision?type=GenVideo.
Virtual Width Networks
Nov 17, 2025



Abstract:We introduce Virtual Width Networks (VWN), a framework that delivers the benefits of wider representations without incurring the quadratic cost of increasing the hidden size. VWN decouples representational width from backbone width, expanding the embedding space while keeping backbone compute nearly constant. In our large-scale experiment, an 8-times expansion accelerates optimization by over 2 times for next-token and 3 times for next-2-token prediction. The advantage amplifies over training as both the loss gap grows and the convergence-speedup ratio increases, showing that VWN is not only token-efficient but also increasingly effective with scale. Moreover, we identify an approximately log-linear scaling relation between virtual width and loss reduction, offering an initial empirical basis and motivation for exploring virtual-width scaling as a new dimension of large-model efficiency.
Depth Anything 3: Recovering the Visual Space from Any Views
Nov 13, 2025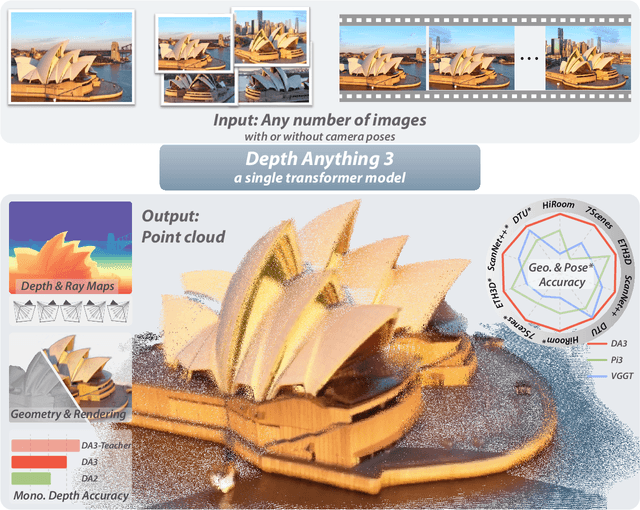
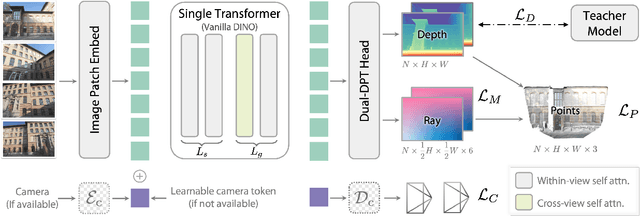
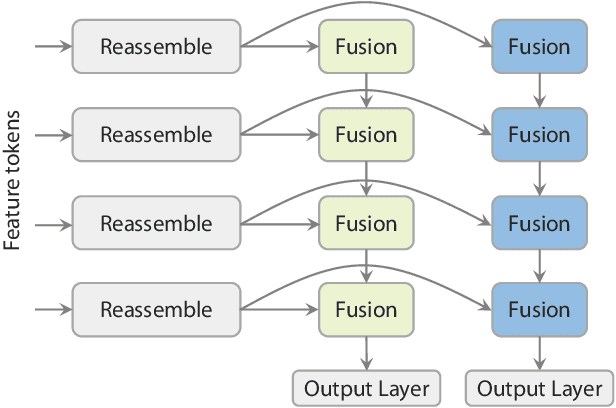
Abstract:We present Depth Anything 3 (DA3), a model that predicts spatially consistent geometry from an arbitrary number of visual inputs, with or without known camera poses. In pursuit of minimal modeling, DA3 yields two key insights: a single plain transformer (e.g., vanilla DINO encoder) is sufficient as a backbone without architectural specialization, and a singular depth-ray prediction target obviates the need for complex multi-task learning. Through our teacher-student training paradigm, the model achieves a level of detail and generalization on par with Depth Anything 2 (DA2). We establish a new visual geometry benchmark covering camera pose estimation, any-view geometry and visual rendering. On this benchmark, DA3 sets a new state-of-the-art across all tasks, surpassing prior SOTA VGGT by an average of 44.3% in camera pose accuracy and 25.1% in geometric accuracy. Moreover, it outperforms DA2 in monocular depth estimation. All models are trained exclusively on public academic datasets.
Lumine: An Open Recipe for Building Generalist Agents in 3D Open Worlds
Nov 12, 2025
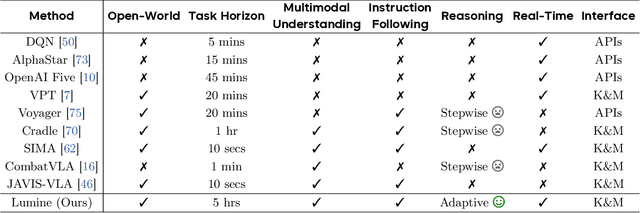

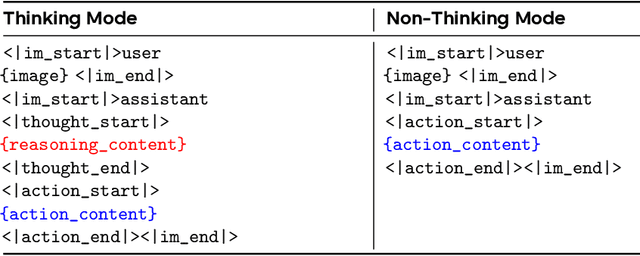
Abstract:We introduce Lumine, the first open recipe for developing generalist agents capable of completing hours-long complex missions in real time within challenging 3D open-world environments. Lumine adopts a human-like interaction paradigm that unifies perception, reasoning, and action in an end-to-end manner, powered by a vision-language model. It processes raw pixels at 5 Hz to produce precise 30 Hz keyboard-mouse actions and adaptively invokes reasoning only when necessary. Trained in Genshin Impact, Lumine successfully completes the entire five-hour Mondstadt main storyline on par with human-level efficiency and follows natural language instructions to perform a broad spectrum of tasks in both 3D open-world exploration and 2D GUI manipulation across collection, combat, puzzle-solving, and NPC interaction. In addition to its in-domain performance, Lumine demonstrates strong zero-shot cross-game generalization. Without any fine-tuning, it accomplishes 100-minute missions in Wuthering Waves and the full five-hour first chapter of Honkai: Star Rail. These promising results highlight Lumine's effectiveness across distinct worlds and interaction dynamics, marking a concrete step toward generalist agents in open-ended environments.
Understanding Transformer from the Perspective of Associative Memory
May 26, 2025Abstract:In this paper, we share our reflections and insights on understanding Transformer architectures through the lens of associative memory--a classic psychological concept inspired by human cognition. We start with the basics of associative memory (think simple linear attention) and then dive into two dimensions: Memory Capacity: How much can a Transformer really remember, and how well? We introduce retrieval SNR to measure this and use a kernel perspective to mathematically reveal why Softmax Attention is so effective. We also show how FFNs can be seen as a type of associative memory, leading to insights on their design and potential improvements. Memory Update: How do these memories learn and evolve? We present a unified framework for understanding how different Transformer variants (like DeltaNet and Softmax Attention) update their "knowledge base". This leads us to tackle two provocative questions: 1. Are Transformers fundamentally limited in what they can express, and can we break these barriers? 2. If a Transformer had infinite context, would it become infinitely intelligent? We want to demystify Transformer architecture, offering a clearer understanding of existing designs. This exploration aims to provide fresh insights and spark new avenues for Transformer innovation.
Emerging Properties in Unified Multimodal Pretraining
May 20, 2025



Abstract:Unifying multimodal understanding and generation has shown impressive capabilities in cutting-edge proprietary systems. In this work, we introduce BAGEL, an open0source foundational model that natively supports multimodal understanding and generation. BAGEL is a unified, decoder0only model pretrained on trillions of tokens curated from large0scale interleaved text, image, video, and web data. When scaled with such diverse multimodal interleaved data, BAGEL exhibits emerging capabilities in complex multimodal reasoning. As a result, it significantly outperforms open-source unified models in both multimodal generation and understanding across standard benchmarks, while exhibiting advanced multimodal reasoning abilities such as free-form image manipulation, future frame prediction, 3D manipulation, and world navigation. In the hope of facilitating further opportunities for multimodal research, we share the key findings, pretraining details, data creation protocal, and release our code and checkpoints to the community. The project page is at https://bagel-ai.org/
Seed1.5-VL Technical Report
May 11, 2025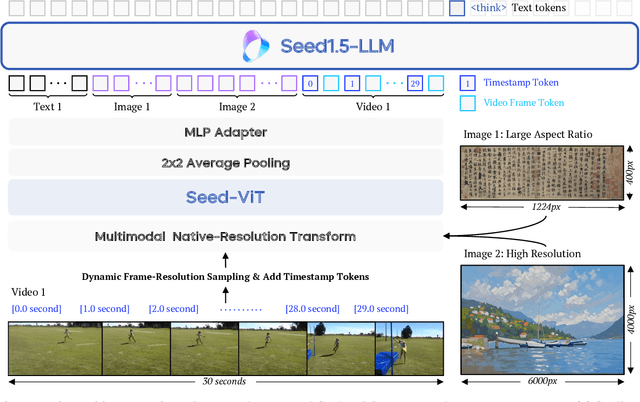

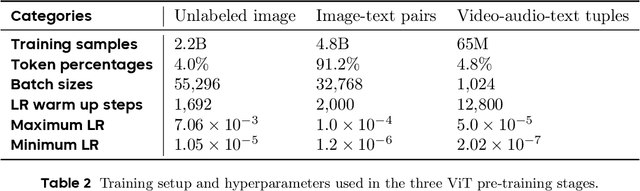
Abstract:We present Seed1.5-VL, a vision-language foundation model designed to advance general-purpose multimodal understanding and reasoning. Seed1.5-VL is composed with a 532M-parameter vision encoder and a Mixture-of-Experts (MoE) LLM of 20B active parameters. Despite its relatively compact architecture, it delivers strong performance across a wide spectrum of public VLM benchmarks and internal evaluation suites, achieving the state-of-the-art performance on 38 out of 60 public benchmarks. Moreover, in agent-centric tasks such as GUI control and gameplay, Seed1.5-VL outperforms leading multimodal systems, including OpenAI CUA and Claude 3.7. Beyond visual and video understanding, it also demonstrates strong reasoning abilities, making it particularly effective for multimodal reasoning challenges such as visual puzzles. We believe these capabilities will empower broader applications across diverse tasks. In this report, we mainly provide a comprehensive review of our experiences in building Seed1.5-VL across model design, data construction, and training at various stages, hoping that this report can inspire further research. Seed1.5-VL is now accessible at https://www.volcengine.com/ (Volcano Engine Model ID: doubao-1-5-thinking-vision-pro-250428)
UI-TARS: Pioneering Automated GUI Interaction with Native Agents
Jan 21, 2025Abstract:This paper introduces UI-TARS, a native GUI agent model that solely perceives the screenshots as input and performs human-like interactions (e.g., keyboard and mouse operations). Unlike prevailing agent frameworks that depend on heavily wrapped commercial models (e.g., GPT-4o) with expert-crafted prompts and workflows, UI-TARS is an end-to-end model that outperforms these sophisticated frameworks. Experiments demonstrate its superior performance: UI-TARS achieves SOTA performance in 10+ GUI agent benchmarks evaluating perception, grounding, and GUI task execution. Notably, in the OSWorld benchmark, UI-TARS achieves scores of 24.6 with 50 steps and 22.7 with 15 steps, outperforming Claude (22.0 and 14.9 respectively). In AndroidWorld, UI-TARS achieves 46.6, surpassing GPT-4o (34.5). UI-TARS incorporates several key innovations: (1) Enhanced Perception: leveraging a large-scale dataset of GUI screenshots for context-aware understanding of UI elements and precise captioning; (2) Unified Action Modeling, which standardizes actions into a unified space across platforms and achieves precise grounding and interaction through large-scale action traces; (3) System-2 Reasoning, which incorporates deliberate reasoning into multi-step decision making, involving multiple reasoning patterns such as task decomposition, reflection thinking, milestone recognition, etc. (4) Iterative Training with Reflective Online Traces, which addresses the data bottleneck by automatically collecting, filtering, and reflectively refining new interaction traces on hundreds of virtual machines. Through iterative training and reflection tuning, UI-TARS continuously learns from its mistakes and adapts to unforeseen situations with minimal human intervention. We also analyze the evolution path of GUI agents to guide the further development of this domain.
Cancer Subtyping by Improved Transcriptomic Features Using Vector Quantized Variational Autoencoder
Jul 20, 2022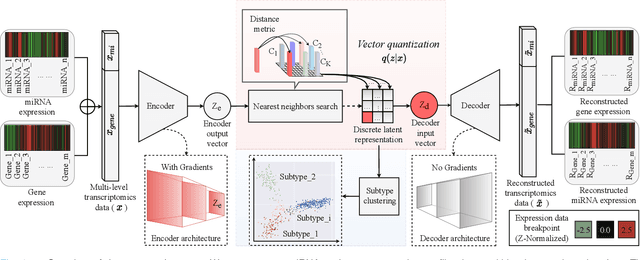
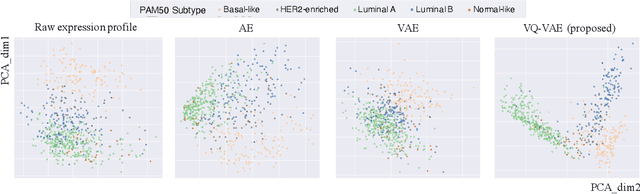
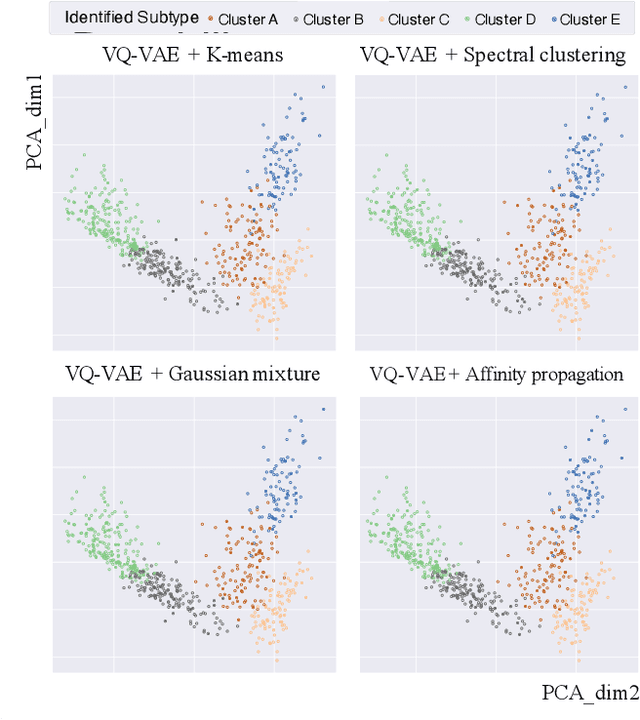
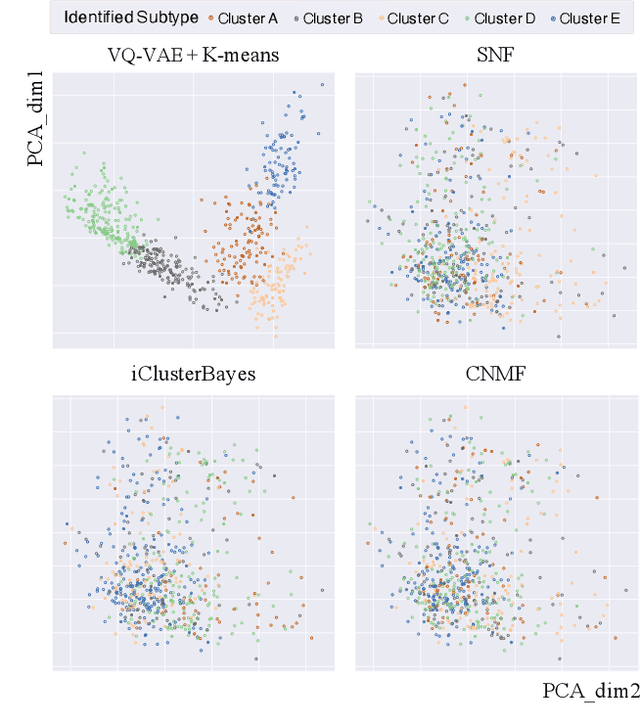
Abstract:Defining and separating cancer subtypes is essential for facilitating personalized therapy modality and prognosis of patients. The definition of subtypes has been constantly recalibrated as a result of our deepened understanding. During this recalibration, researchers often rely on clustering of cancer data to provide an intuitive visual reference that could reveal the intrinsic characteristics of subtypes. The data being clustered are often omics data such as transcriptomics that have strong correlations to the underlying biological mechanism. However, while existing studies have shown promising results, they suffer from issues associated with omics data: sample scarcity and high dimensionality. As such, existing methods often impose unrealistic assumptions to extract useful features from the data while avoiding overfitting to spurious correlations. In this paper, we propose to leverage a recent strong generative model, Vector Quantized Variational AutoEncoder (VQ-VAE), to tackle the data issues and extract informative latent features that are crucial to the quality of subsequent clustering by retaining only information relevant to reconstructing the input. VQ-VAE does not impose strict assumptions and hence its latent features are better representations of the input, capable of yielding superior clustering performance with any mainstream clustering method. Extensive experiments and medical analysis on multiple datasets comprising 10 distinct cancers demonstrate the VQ-VAE clustering results can significantly and robustly improve prognosis over prevalent subtyping systems.
Adaptive Spike-Like Representation of EEG Signals for Sleep Stages Scoring
Apr 02, 2022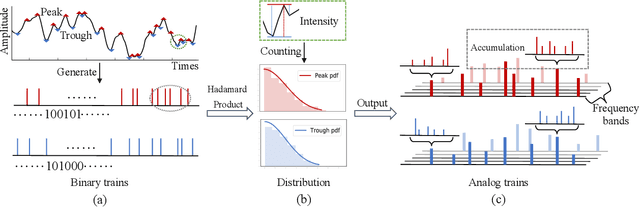
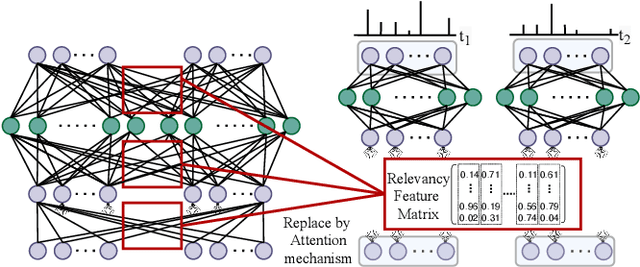
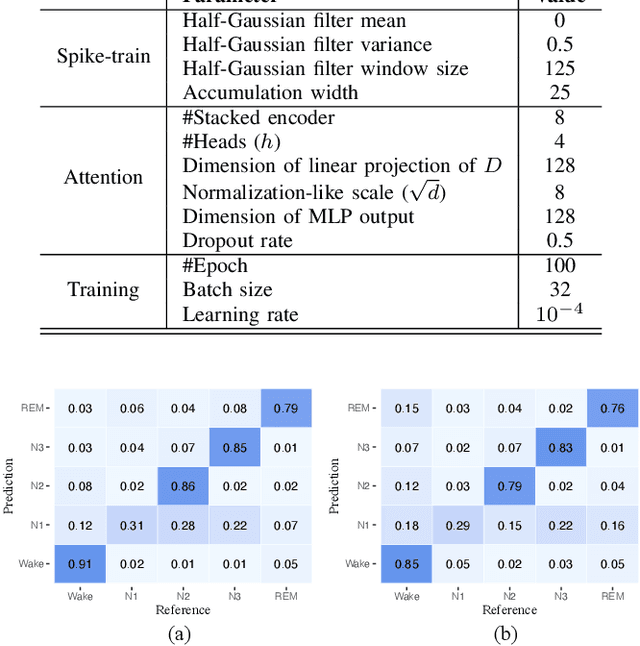
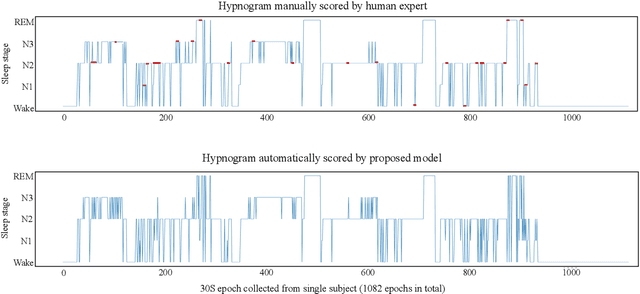
Abstract:Recently there has seen promising results on automatic stage scoring by extracting spatio-temporal features from electroencephalogram (EEG). Such methods entail laborious manual feature engineering and domain knowledge. In this study, we propose an adaptive scheme to probabilistically encode, filter and accumulate the input signals and weight the resultant features by the half-Gaussian probabilities of signal intensities. The adaptive representations are subsequently fed into a transformer model to automatically mine the relevance between features and corresponding stages. Extensive experiments on the largest public dataset against state-of-the-art methods validate the effectiveness of our proposed method and reveal promising future directions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge