Kuan-Jui Su
PF-DAformer: Proximal Femur Segmentation via Domain Adaptive Transformer for Dual-Center QCT
Oct 30, 2025Abstract:Quantitative computed tomography (QCT) plays a crucial role in assessing bone strength and fracture risk by enabling volumetric analysis of bone density distribution in the proximal femur. However, deploying automated segmentation models in practice remains difficult because deep networks trained on one dataset often fail when applied to another. This failure stems from domain shift, where scanners, reconstruction settings, and patient demographics vary across institutions, leading to unstable predictions and unreliable quantitative metrics. Overcoming this barrier is essential for multi-center osteoporosis research and for ensuring that radiomics and structural finite element analysis results remain reproducible across sites. In this work, we developed a domain-adaptive transformer segmentation framework tailored for multi-institutional QCT. Our model is trained and validated on one of the largest hip fracture related research cohorts to date, comprising 1,024 QCT images scans from Tulane University and 384 scans from Rochester, Minnesota for proximal femur segmentation. To address domain shift, we integrate two complementary strategies within a 3D TransUNet backbone: adversarial alignment via Gradient Reversal Layer (GRL), which discourages the network from encoding site-specific cues, and statistical alignment via Maximum Mean Discrepancy (MMD), which explicitly reduces distributional mismatches between institutions. This dual mechanism balances invariance and fine-grained alignment, enabling scanner-agnostic feature learning while preserving anatomical detail.
ICGM-FRAX: Iterative Cross Graph Matching for Hip Fracture Risk Assessment using Dual-energy X-ray Absorptiometry Images
Apr 21, 2025Abstract:Hip fractures represent a major health concern, particularly among the elderly, often leading decreased mobility and increased mortality. Early and accurate detection of at risk individuals is crucial for effective intervention. In this study, we propose Iterative Cross Graph Matching for Hip Fracture Risk Assessment (ICGM-FRAX), a novel approach for predicting hip fractures using Dual-energy X-ray Absorptiometry (DXA) images. ICGM-FRAX involves iteratively comparing a test (subject) graph with multiple template graphs representing the characteristics of hip fracture subjects to assess the similarity and accurately to predict hip fracture risk. These graphs are obtained as follows. The DXA images are separated into multiple regions of interest (RoIs), such as the femoral head, shaft, and lesser trochanter. Radiomic features are then calculated for each RoI, with the central coordinates used as nodes in a graph. The connectivity between nodes is established according to the Euclidean distance between these coordinates. This process transforms each DXA image into a graph, where each node represents a RoI, and edges derived by the centroids of RoIs capture the spatial relationships between them. If the test graph closely matches a set of template graphs representing subjects with incident hip fractures, it is classified as indicating high hip fracture risk. We evaluated our method using 547 subjects from the UK Biobank dataset, and experimental results show that ICGM-FRAX achieved a sensitivity of 0.9869, demonstrating high accuracy in predicting hip fractures.
A Privacy-Preserving Domain Adversarial Federated learning for multi-site brain functional connectivity analysis
Feb 03, 2025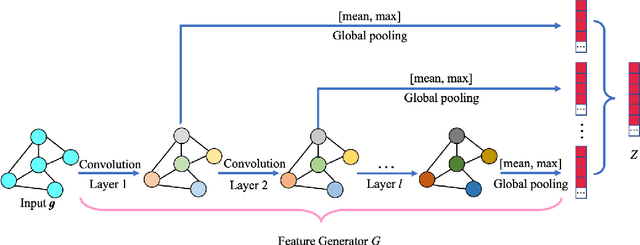
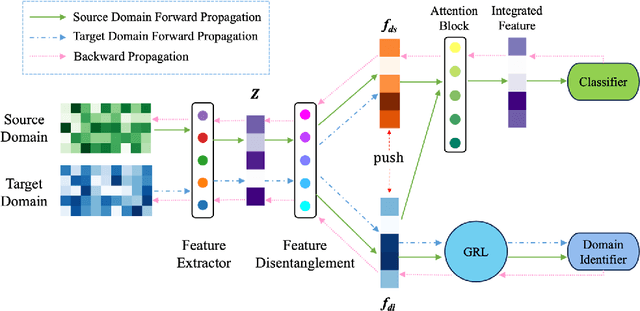
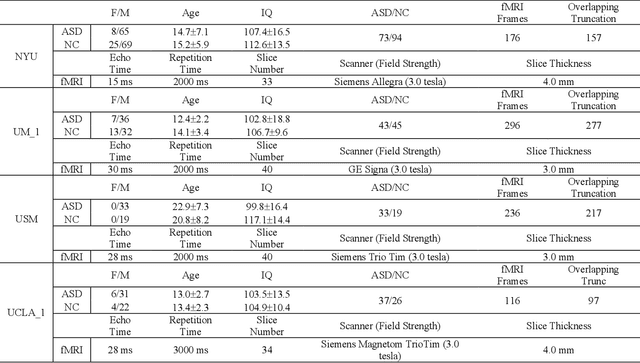
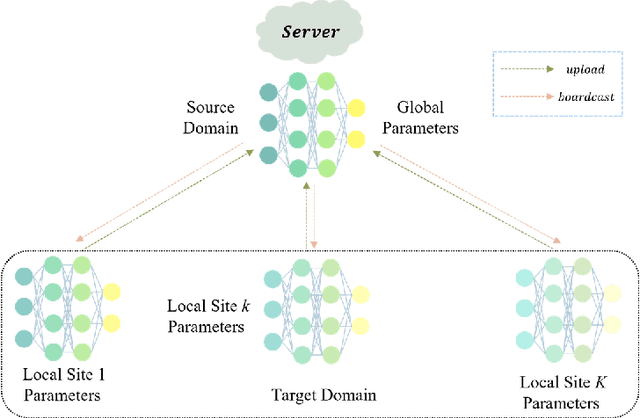
Abstract:Resting-state functional magnetic resonance imaging (rs-fMRI) and its derived functional connectivity networks (FCNs) have become critical for understanding neurological disorders. However, collaborative analyses and the generalizability of models still face significant challenges due to privacy regulations and the non-IID (non-independent and identically distributed) property of multiple data sources. To mitigate these difficulties, we propose Domain Adversarial Federated Learning (DAFed), a novel federated deep learning framework specifically designed for non-IID fMRI data analysis in multi-site settings. DAFed addresses these challenges through feature disentanglement, decomposing the latent feature space into domain-invariant and domain-specific components, to ensure robust global learning while preserving local data specificity. Furthermore, adversarial training facilitates effective knowledge transfer between labeled and unlabeled datasets, while a contrastive learning module enhances the global representation of domain-invariant features. We evaluated DAFed on the diagnosis of ASD and further validated its generalizability in the classification of AD, demonstrating its superior classification accuracy compared to state-of-the-art methods. Additionally, an enhanced Score-CAM module identifies key brain regions and functional connectivity significantly associated with ASD and MCI, respectively, uncovering shared neurobiological patterns across sites. These findings highlight the potential of DAFed to advance multi-site collaborative research in neuroimaging while protecting data confidentiality.
A Staged Approach using Machine Learning and Uncertainty Quantification to Predict the Risk of Hip Fracture
May 30, 2024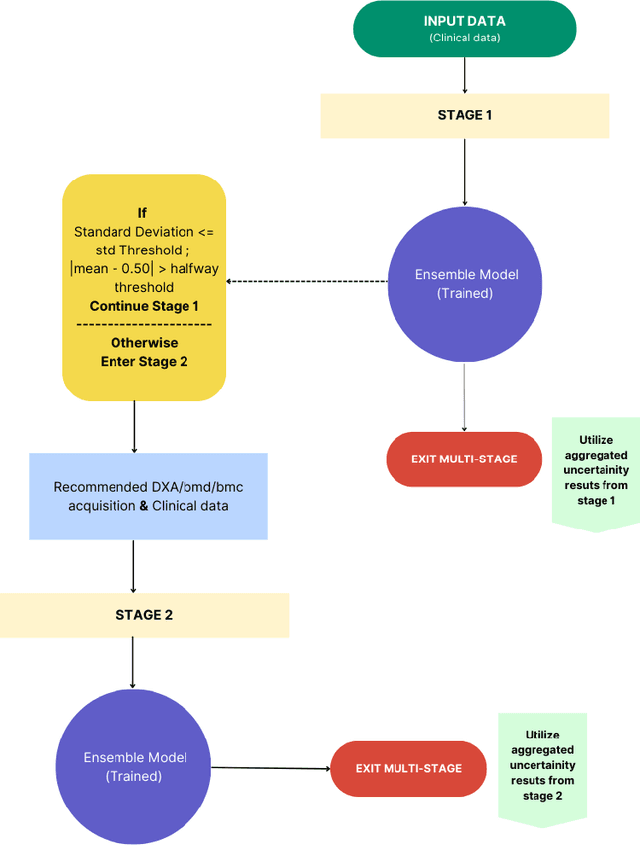
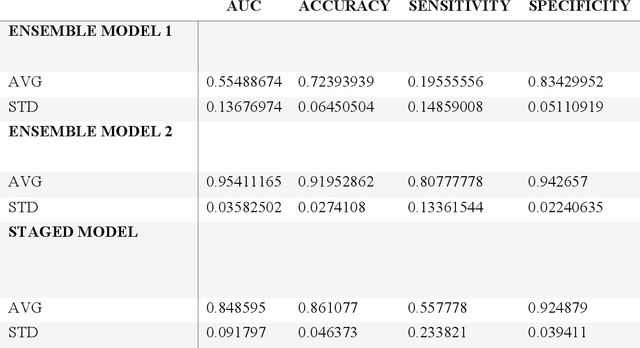
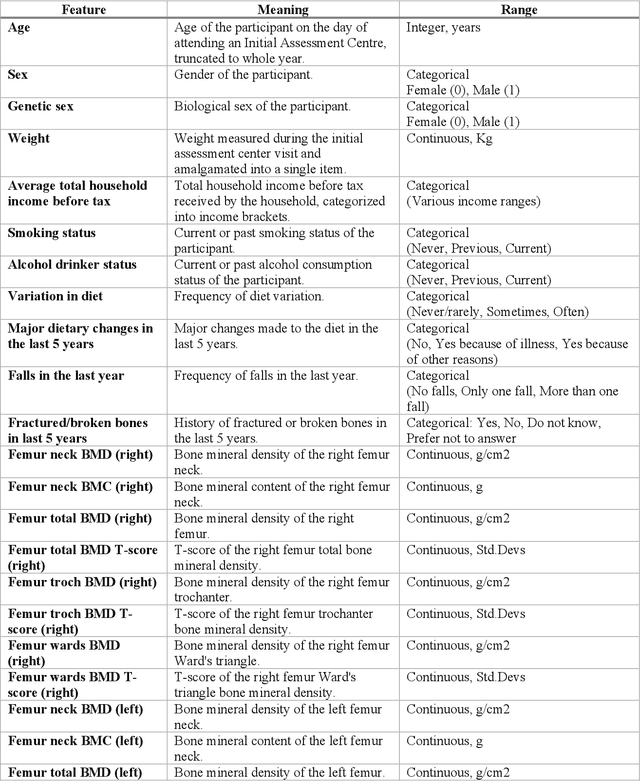
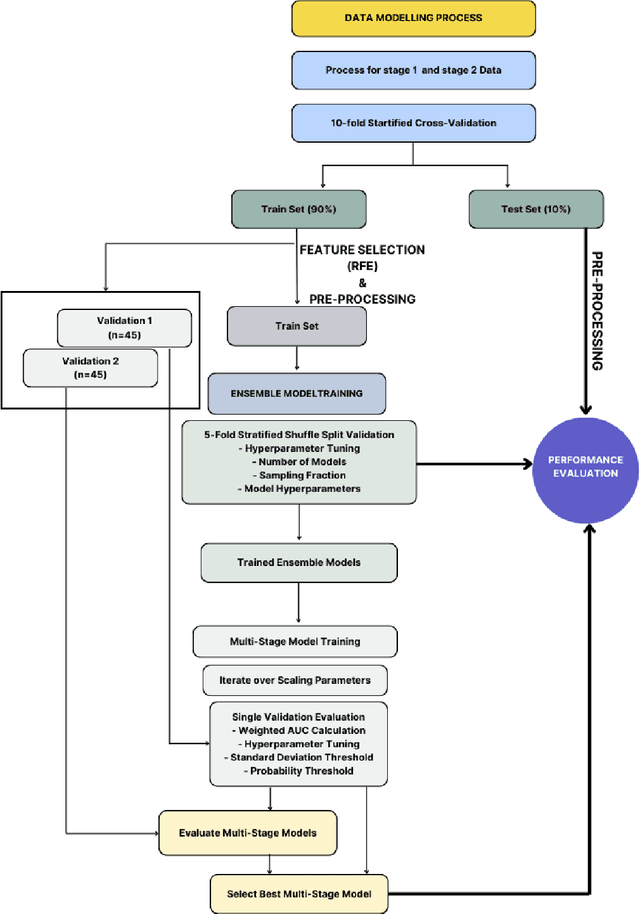
Abstract:Despite advancements in medical care, hip fractures impose a significant burden on individuals and healthcare systems. This paper focuses on the prediction of hip fracture risk in older and middle-aged adults, where falls and compromised bone quality are predominant factors. We propose a novel staged model that combines advanced imaging and clinical data to improve predictive performance. By using CNNs to extract features from hip DXA images, along with clinical variables, shape measurements, and texture features, our method provides a comprehensive framework for assessing fracture risk. A staged machine learning-based model was developed using two ensemble models: Ensemble 1 (clinical variables only) and Ensemble 2 (clinical variables and DXA imaging features). This staged approach used uncertainty quantification from Ensemble 1 to decide if DXA features are necessary for further prediction. Ensemble 2 exhibited the highest performance, achieving an AUC of 0.9541, an accuracy of 0.9195, a sensitivity of 0.8078, and a specificity of 0.9427. The staged model also performed well, with an AUC of 0.8486, an accuracy of 0.8611, a sensitivity of 0.5578, and a specificity of 0.9249, outperforming Ensemble 1, which had an AUC of 0.5549, an accuracy of 0.7239, a sensitivity of 0.1956, and a specificity of 0.8343. Furthermore, the staged model suggested that 54.49% of patients did not require DXA scanning. It effectively balanced accuracy and specificity, offering a robust solution when DXA data acquisition is not always feasible. Statistical tests confirmed significant differences between the models, highlighting the advantages of the advanced modeling strategies. Our staged approach could identify individuals at risk with a high accuracy but reduce the unnecessary DXA scanning. It has great promise to guide interventions to prevent hip fractures with reduced cost and radiation.
Multi-View Variational Autoencoder for Missing Value Imputation in Untargeted Metabolomics
Oct 12, 2023


Abstract:Background: Missing data is a common challenge in mass spectrometry-based metabolomics, which can lead to biased and incomplete analyses. The integration of whole-genome sequencing (WGS) data with metabolomics data has emerged as a promising approach to enhance the accuracy of data imputation in metabolomics studies. Method: In this study, we propose a novel method that leverages the information from WGS data and reference metabolites to impute unknown metabolites. Our approach utilizes a multi-view variational autoencoder to jointly model the burden score, polygenetic risk score (PGS), and linkage disequilibrium (LD) pruned single nucleotide polymorphisms (SNPs) for feature extraction and missing metabolomics data imputation. By learning the latent representations of both omics data, our method can effectively impute missing metabolomics values based on genomic information. Results: We evaluate the performance of our method on empirical metabolomics datasets with missing values and demonstrate its superiority compared to conventional imputation techniques. Using 35 template metabolites derived burden scores, PGS and LD-pruned SNPs, the proposed methods achieved r2-scores > 0.01 for 71.55% of metabolites. Conclusion: The integration of WGS data in metabolomics imputation not only improves data completeness but also enhances downstream analyses, paving the way for more comprehensive and accurate investigations of metabolic pathways and disease associations. Our findings offer valuable insights into the potential benefits of utilizing WGS data for metabolomics data imputation and underscore the importance of leveraging multi-modal data integration in precision medicine research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge