Tian Qin
Drawback of Enforcing Equivariance and its Compensation via the Lens of Expressive Power
Dec 10, 2025Abstract:Equivariant neural networks encode symmetry as an inductive bias and have achieved strong empirical performance in wide domains. However, their expressive power remains not well understood. Focusing on 2-layer ReLU networks, this paper investigates the impact of equivariance constraints on the expressivity of equivariant and layer-wise equivariant networks. By examining the boundary hyperplanes and the channel vectors of ReLU networks, we construct an example showing that equivariance constraints could strictly limit expressive power. However, we demonstrate that this drawback can be compensated via enlarging the model size. Furthermore, we show that despite a larger model size, the resulting architecture could still correspond to a hypothesis space with lower complexity, implying superior generalizability for equivariant networks.
BootOOD: Self-Supervised Out-of-Distribution Detection via Synthetic Sample Exposure under Neural Collapse
Nov 17, 2025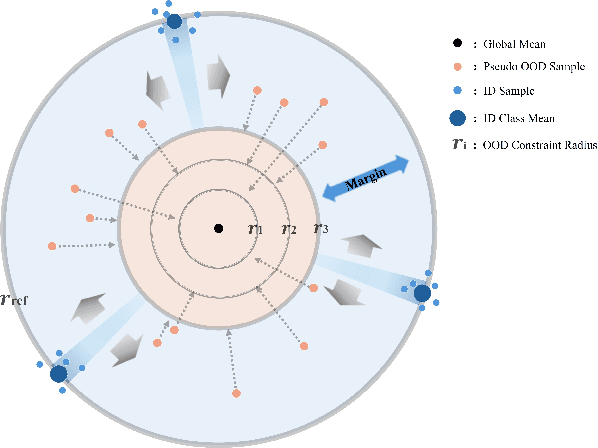
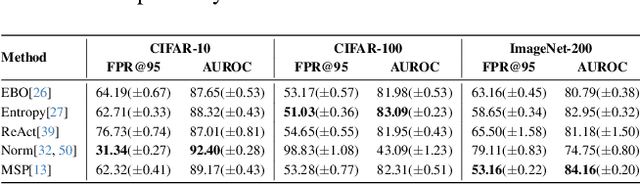
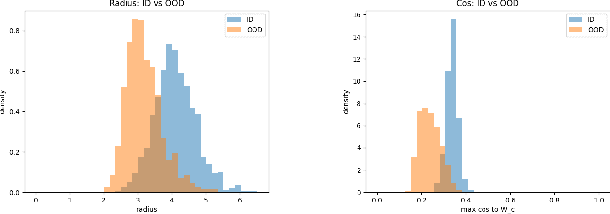
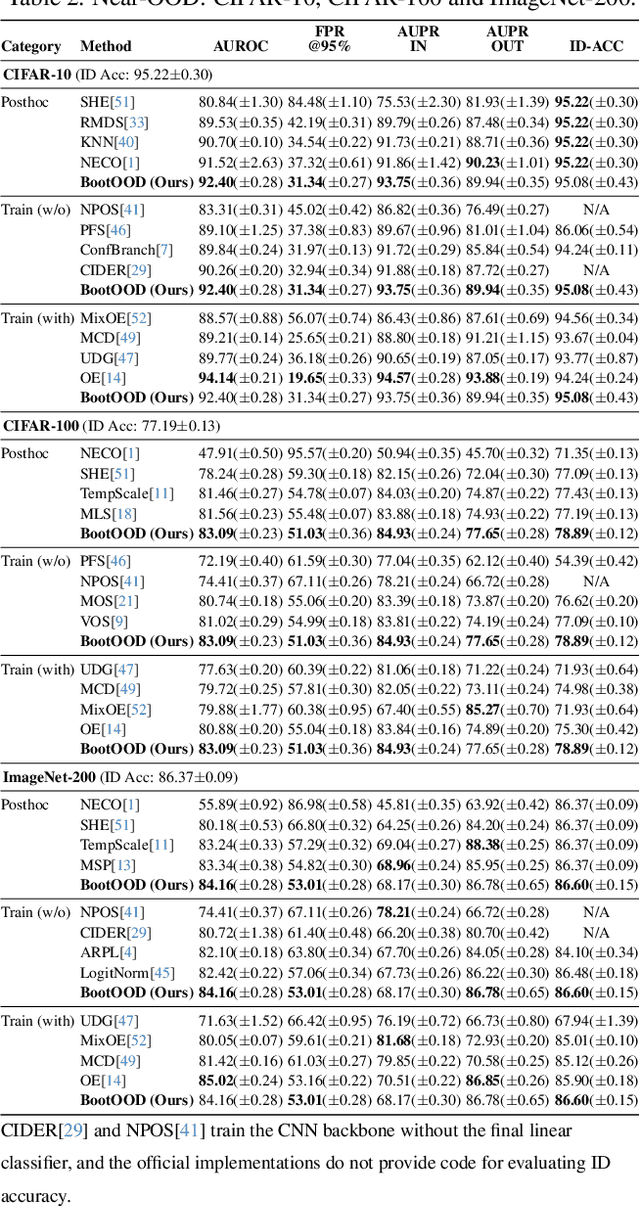
Abstract:Out-of-distribution (OOD) detection is critical for deploying image classifiers in safety-sensitive environments, yet existing detectors often struggle when OOD samples are semantically similar to the in-distribution (ID) classes. We present BootOOD, a fully self-supervised OOD detection framework that bootstraps exclusively from ID data and is explicitly designed to handle semantically challenging OOD samples. BootOOD synthesizes pseudo-OOD features through simple transformations of ID representations and leverages Neural Collapse (NC), where ID features cluster tightly around class means with consistent feature norms. Unlike prior approaches that aim to constrain OOD features into subspaces orthogonal to the collapsed ID means, BootOOD introduces a lightweight auxiliary head that performs radius-based classification on feature norms. This design decouples OOD detection from the primary classifier and imposes a relaxed requirement: OOD samples are learned to have smaller feature norms than ID features, which is easier to satisfy when ID and OOD are semantically close. Experiments on CIFAR-10, CIFAR-100, and ImageNet-200 show that BootOOD outperforms prior post-hoc methods, surpasses training-based methods without outlier exposure, and is competitive with state-of-the-art outlier-exposure approaches while maintaining or improving ID accuracy.
COMPASS: A Multi-Turn Benchmark for Tool-Mediated Planning & Preference Optimization
Oct 08, 2025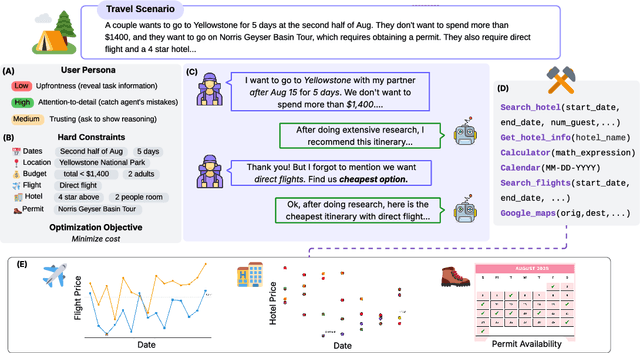
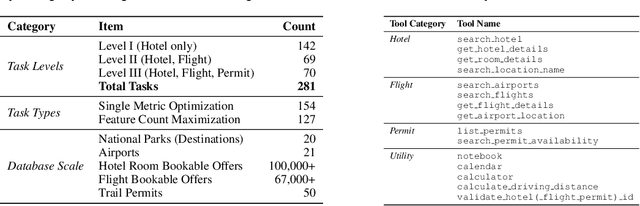
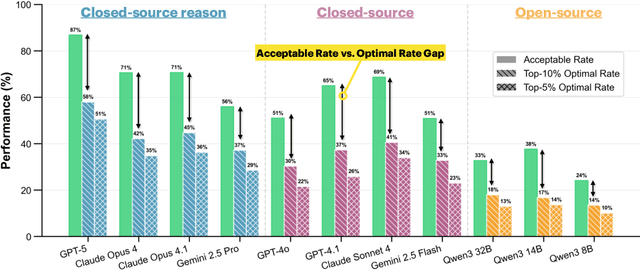
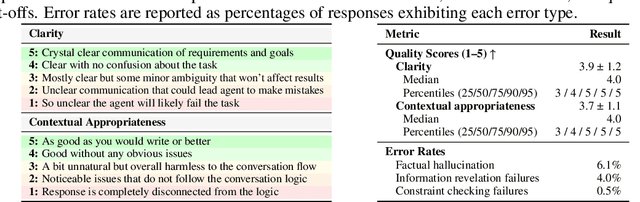
Abstract:Real-world large language model (LLM) agents must master strategic tool use and user preference optimization through multi-turn interactions to assist users with complex planning tasks. We introduce COMPASS (Constrained Optimization through Multi-turn Planning and Strategic Solutions), a benchmark that evaluates agents on realistic travel-planning scenarios. We cast travel planning as a constrained preference optimization problem, where agents must satisfy hard constraints while simultaneously optimizing soft user preferences. To support this, we build a realistic travel database covering transportation, accommodation, and ticketing for 20 U.S. National Parks, along with a comprehensive tool ecosystem that mirrors commercial booking platforms. Evaluating state-of-the-art models, we uncover two critical gaps: (i) an acceptable-optimal gap, where agents reliably meet constraints but fail to optimize preferences, and (ii) a plan-coordination gap, where performance collapses on multi-service (flight and hotel) coordination tasks, especially for open-source models. By grounding reasoning and planning in a practical, user-facing domain, COMPASS provides a benchmark that directly measures an agent's ability to optimize user preferences in realistic tasks, bridging theoretical advances with real-world impact.
From Static to Adaptive Defense: Federated Multi-Agent Deep Reinforcement Learning-Driven Moving Target Defense Against DoS Attacks in UAV Swarm Networks
Jun 09, 2025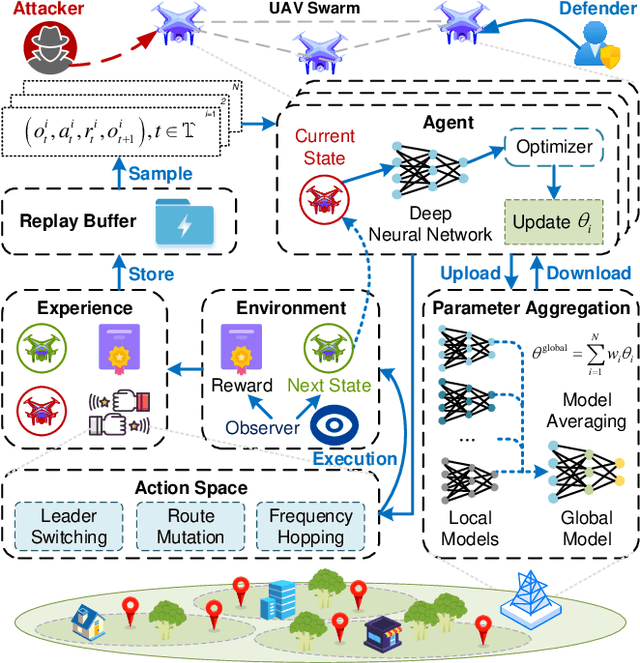
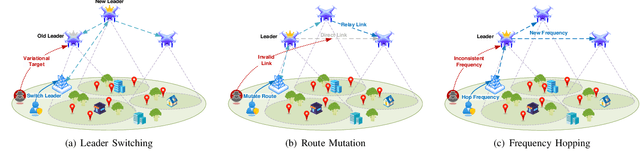
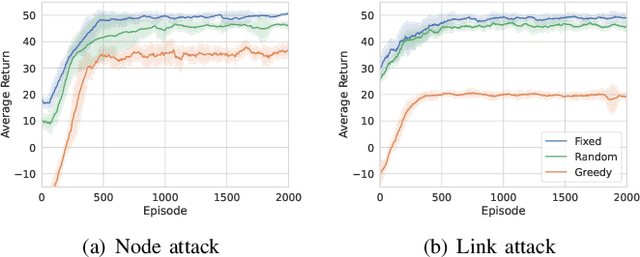
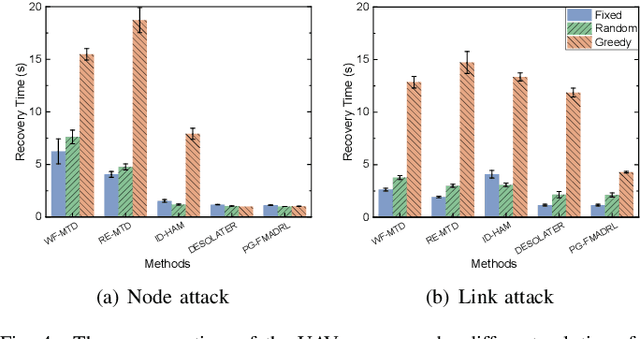
Abstract:The proliferation of unmanned aerial vehicle (UAV) swarms has enabled a wide range of mission-critical applications, but also exposes UAV networks to severe Denial-of-Service (DoS) threats due to their open wireless environment, dynamic topology, and resource constraints. Traditional static or centralized defense mechanisms are often inadequate for such dynamic and distributed scenarios. To address these challenges, we propose a novel federated multi-agent deep reinforcement learning (FMADRL)-driven moving target defense (MTD) framework for proactive and adaptive DoS mitigation in UAV swarm networks. Specifically, we design three lightweight and coordinated MTD mechanisms, including leader switching, route mutation, and frequency hopping, that leverage the inherent flexibility of UAV swarms to disrupt attacker efforts and enhance network resilience. The defense problem is formulated as a multi-agent partially observable Markov decision process (POMDP), capturing the distributed, resource-constrained, and uncertain nature of UAV swarms under attack. Each UAV is equipped with a local policy agent that autonomously selects MTD actions based on partial observations and local experiences. By employing a policy gradient-based FMADRL algorithm, UAVs collaboratively optimize their defense policies via reward-weighted aggregation, enabling distributed learning without sharing raw data and thus reducing communication overhead. Extensive simulations demonstrate that our approach significantly outperforms state-of-the-art baselines, achieving up to a 34.6% improvement in attack mitigation rate, a reduction in average recovery time of up to 94.6%, and decreases in energy consumption and defense cost by as much as 29.3% and 98.3%, respectively, while maintaining robust mission continuity under various DoS attack strategies.
Decomposing Elements of Problem Solving: What "Math" Does RL Teach?
May 28, 2025Abstract:Mathematical reasoning tasks have become prominent benchmarks for assessing the reasoning capabilities of LLMs, especially with reinforcement learning (RL) methods such as GRPO showing significant performance gains. However, accuracy metrics alone do not support fine-grained assessment of capabilities and fail to reveal which problem-solving skills have been internalized. To better understand these capabilities, we propose to decompose problem solving into fundamental capabilities: Plan (mapping questions to sequences of steps), Execute (correctly performing solution steps), and Verify (identifying the correctness of a solution). Empirically, we find that GRPO mainly enhances the execution skill-improving execution robustness on problems the model already knows how to solve-a phenomenon we call temperature distillation. More importantly, we show that RL-trained models struggle with fundamentally new problems, hitting a 'coverage wall' due to insufficient planning skills. To explore RL's impact more deeply, we construct a minimal, synthetic solution-tree navigation task as an analogy for mathematical problem-solving. This controlled setup replicates our empirical findings, confirming RL primarily boosts execution robustness. Importantly, in this setting, we identify conditions under which RL can potentially overcome the coverage wall through improved exploration and generalization to new solution paths. Our findings provide insights into the role of RL in enhancing LLM reasoning, expose key limitations, and suggest a path toward overcoming these barriers. Code is available at https://github.com/cfpark00/RL-Wall.
To Backtrack or Not to Backtrack: When Sequential Search Limits Model Reasoning
Apr 09, 2025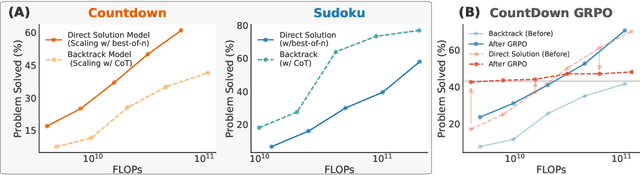

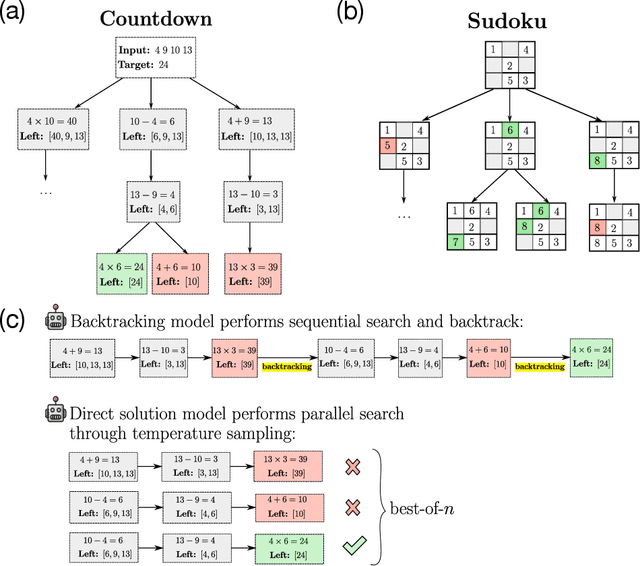
Abstract:Recent advancements in large language models have significantly improved their reasoning abilities, particularly through techniques involving search and backtracking. Backtracking naturally scales test-time compute by enabling sequential, linearized exploration via long chain-of-thought (CoT) generation. However, this is not the only strategy for scaling test-time compute: parallel sampling with best-of-n selection provides an alternative that generates diverse solutions simultaneously. Despite the growing adoption of sequential search, its advantages over parallel sampling--especially under a fixed compute budget remain poorly understood. In this paper, we systematically compare these two approaches on two challenging reasoning tasks: CountDown and Sudoku. Surprisingly, we find that sequential search underperforms parallel sampling on CountDown but outperforms it on Sudoku, suggesting that backtracking is not universally beneficial. We identify two factors that can cause backtracking to degrade performance: (1) training on fixed search traces can lock models into suboptimal strategies, and (2) explicit CoT supervision can discourage "implicit" (non-verbalized) reasoning. Extending our analysis to reinforcement learning (RL), we show that models with backtracking capabilities benefit significantly from RL fine-tuning, while models without backtracking see limited, mixed gains. Together, these findings challenge the assumption that backtracking universally enhances LLM reasoning, instead revealing a complex interaction between task structure, training data, model scale, and learning paradigm.
Debiasing Kernel-Based Generative Models
Mar 26, 2025Abstract:We propose a novel two-stage framework of generative models named Debiasing Kernel-Based Generative Models (DKGM) with the insights from kernel density estimation (KDE) and stochastic approximation. In the first stage of DKGM, we employ KDE to bypass the obstacles in estimating the density of data without losing too much image quality. One characteristic of KDE is oversmoothing, which makes the generated image blurry. Therefore, in the second stage, we formulate the process of reducing the blurriness of images as a statistical debiasing problem and develop a novel iterative algorithm to improve image quality, which is inspired by the stochastic approximation. Extensive experiments illustrate that the image quality of DKGM on CIFAR10 is comparable to state-of-the-art models such as diffusion models and GAN models. The performance of DKGM on CelebA 128x128 and LSUN (Church) 128x128 is also competitive. We conduct extra experiments to exploit how the bandwidth in KDE affects the sample diversity and debiasing effect of DKGM. The connections between DKGM and score-based models are also discussed.
Distributional Scaling Laws for Emergent Capabilities
Feb 24, 2025Abstract:In this paper, we explore the nature of sudden breakthroughs in language model performance at scale, which stands in contrast to smooth improvements governed by scaling laws. While advocates of "emergence" view abrupt performance gains as capabilities unlocking at specific scales, others have suggested that they are produced by thresholding effects and alleviated by continuous metrics. We propose that breakthroughs are instead driven by continuous changes in the probability distribution of training outcomes, particularly when performance is bimodally distributed across random seeds. In synthetic length generalization tasks, we show that different random seeds can produce either highly linear or emergent scaling trends. We reveal that sharp breakthroughs in metrics are produced by underlying continuous changes in their distribution across seeds. Furthermore, we provide a case study of inverse scaling and show that even as the probability of a successful run declines, the average performance of a successful run continues to increase monotonically. We validate our distributional scaling framework on realistic settings by measuring MMLU performance in LLM populations. These insights emphasize the role of random variation in the effect of scale on LLM capabilities.
Sometimes I am a Tree: Data Drives Unstable Hierarchical Generalization
Dec 05, 2024Abstract:Neural networks often favor shortcut heuristics based on surface-level patterns. As one example, language models (LMs) behave like n-gram models early in training. However, to correctly apply grammatical rules, LMs must rely on hierarchical syntactic representations instead of n-grams. In this work, we use cases studies of English grammar to explore how latent structure in training data drives models toward improved out-of-distribution (OOD) generalization.We then investigate how data composition can lead to inconsistent OOD behavior across random seeds and to unstable training dynamics. Our results show that models stabilize in their OOD behavior only when they fully commit to either a surface-level linear rule or a hierarchical rule. The hierarchical rule, furthermore, is induced by grammatically complex sequences with deep embedding structures, whereas the linear rule is induced by simpler sequences. When the data contains a mix of simple and complex examples, potential rules compete; each independent training run either stabilizes by committing to a single rule or remains unstable in its OOD behavior. These conditions lead `stable seeds' to cluster around simple rules, forming bimodal performance distributions across seeds. We also identify an exception to the relationship between stability and generalization: models which memorize patterns from low-diversity training data can overfit stably, with different rules for memorized and unmemorized patterns. Our findings emphasize the critical role of training data in shaping generalization patterns and how competition between data subsets contributes to inconsistent generalization outcomes across random seeds. Code is available at https://github.com/sunnytqin/concept_comp.git.
A Label is Worth a Thousand Images in Dataset Distillation
Jun 15, 2024



Abstract:Data $\textit{quality}$ is a crucial factor in the performance of machine learning models, a principle that dataset distillation methods exploit by compressing training datasets into much smaller counterparts that maintain similar downstream performance. Understanding how and why data distillation methods work is vital not only for improving these methods but also for revealing fundamental characteristics of "good" training data. However, a major challenge in achieving this goal is the observation that distillation approaches, which rely on sophisticated but mostly disparate methods to generate synthetic data, have little in common with each other. In this work, we highlight a largely overlooked aspect common to most of these methods: the use of soft (probabilistic) labels. Through a series of ablation experiments, we study the role of soft labels in depth. Our results reveal that the main factor explaining the performance of state-of-the-art distillation methods is not the specific techniques used to generate synthetic data but rather the use of soft labels. Furthermore, we demonstrate that not all soft labels are created equal; they must contain $\textit{structured information}$ to be beneficial. We also provide empirical scaling laws that characterize the effectiveness of soft labels as a function of images-per-class in the distilled dataset and establish an empirical Pareto frontier for data-efficient learning. Combined, our findings challenge conventional wisdom in dataset distillation, underscore the importance of soft labels in learning, and suggest new directions for improving distillation methods. Code for all experiments is available at https://github.com/sunnytqin/no-distillation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge