Vince D. Calhoun
Tri-Institutional Center for Translational Research in Neuroimaging and Data Science
KOCOBrain: Kuramoto-Guided Graph Network for Uncovering Structure-Function Coupling in Adolescent Prenatal Drug Exposure
Jan 16, 2026Abstract:Exposure to psychoactive substances during pregnancy, such as cannabis, can disrupt neurodevelopment and alter large-scale brain networks, yet identifying their neural signatures remains challenging. We introduced KOCOBrain: KuramotO COupled Brain Graph Network; a unified graph neural network framework that integrates structural and functional connectomes via Kuramoto-based phase dynamics and cognition-aware attention. The Kuramoto layer models neural synchronization over anatomical connections, generating phase-informed embeddings that capture structure-function coupling, while cognitive scores modulate information routing in a subject-specific manner followed by a joint objective enhancing robustness under class imbalance scenario. Applied to the ABCD cohort, KOCOBrain improved prenatal drug exposure prediction over relevant baselines and revealed interpretable structure-function patterns that reflect disrupted brain network coordination associated with early exposure.
Deep Deterministic Nonlinear ICA via Total Correlation Minimization with Matrix-Based Entropy Functional
Dec 31, 2025Abstract:Blind source separation, particularly through independent component analysis (ICA), is widely utilized across various signal processing domains for disentangling underlying components from observed mixed signals, owing to its fully data-driven nature that minimizes reliance on prior assumptions. However, conventional ICA methods rely on an assumption of linear mixing, limiting their ability to capture complex nonlinear relationships and to maintain robustness in noisy environments. In this work, we present deep deterministic nonlinear independent component analysis (DDICA), a novel deep neural network-based framework designed to address these limitations. DDICA leverages a matrix-based entropy function to directly optimize the independence criterion via stochastic gradient descent, bypassing the need for variational approximations or adversarial schemes. This results in a streamlined training process and improved resilience to noise. We validated the effectiveness and generalizability of DDICA across a range of applications, including simulated signal mixtures, hyperspectral image unmixing, modeling of primary visual receptive fields, and resting-state functional magnetic resonance imaging (fMRI) data analysis. Experimental results demonstrate that DDICA effectively separates independent components with high accuracy across a range of applications. These findings suggest that DDICA offers a robust and versatile solution for blind source separation in diverse signal processing tasks.
Physics-Guided Multi-View Graph Neural Network for Schizophrenia Classification via Structural-Functional Coupling
May 21, 2025Abstract:Clinical studies reveal disruptions in brain structural connectivity (SC) and functional connectivity (FC) in neuropsychiatric disorders such as schizophrenia (SZ). Traditional approaches might rely solely on SC due to limited functional data availability, hindering comprehension of cognitive and behavioral impairments in individuals with SZ by neglecting the intricate SC-FC interrelationship. To tackle the challenge, we propose a novel physics-guided deep learning framework that leverages a neural oscillation model to describe the dynamics of a collection of interconnected neural oscillators, which operate via nerve fibers dispersed across the brain's structure. Our proposed framework utilizes SC to simultaneously generate FC by learning SC-FC coupling from a system dynamics perspective. Additionally, it employs a novel multi-view graph neural network (GNN) with a joint loss to perform correlation-based SC-FC fusion and classification of individuals with SZ. Experiments conducted on a clinical dataset exhibited improved performance, demonstrating the robustness of our proposed approach.
Unified Cross-Modal Attention-Mixer Based Structural-Functional Connectomics Fusion for Neuropsychiatric Disorder Diagnosis
May 21, 2025Abstract:Gaining insights into the structural and functional mechanisms of the brain has been a longstanding focus in neuroscience research, particularly in the context of understanding and treating neuropsychiatric disorders such as Schizophrenia (SZ). Nevertheless, most of the traditional multimodal deep learning approaches fail to fully leverage the complementary characteristics of structural and functional connectomics data to enhance diagnostic performance. To address this issue, we proposed ConneX, a multimodal fusion method that integrates cross-attention mechanism and multilayer perceptron (MLP)-Mixer for refined feature fusion. Modality-specific backbone graph neural networks (GNNs) were firstly employed to obtain feature representation for each modality. A unified cross-modal attention network was then introduced to fuse these embeddings by capturing intra- and inter-modal interactions, while MLP-Mixer layers refined global and local features, leveraging higher-order dependencies for end-to-end classification with a multi-head joint loss. Extensive evaluations demonstrated improved performance on two distinct clinical datasets, highlighting the robustness of our proposed framework.
MoRE-Brain: Routed Mixture of Experts for Interpretable and Generalizable Cross-Subject fMRI Visual Decoding
May 21, 2025Abstract:Decoding visual experiences from fMRI offers a powerful avenue to understand human perception and develop advanced brain-computer interfaces. However, current progress often prioritizes maximizing reconstruction fidelity while overlooking interpretability, an essential aspect for deriving neuroscientific insight. To address this gap, we propose MoRE-Brain, a neuro-inspired framework designed for high-fidelity, adaptable, and interpretable visual reconstruction. MoRE-Brain uniquely employs a hierarchical Mixture-of-Experts architecture where distinct experts process fMRI signals from functionally related voxel groups, mimicking specialized brain networks. The experts are first trained to encode fMRI into the frozen CLIP space. A finetuned diffusion model then synthesizes images, guided by expert outputs through a novel dual-stage routing mechanism that dynamically weighs expert contributions across the diffusion process. MoRE-Brain offers three main advancements: First, it introduces a novel Mixture-of-Experts architecture grounded in brain network principles for neuro-decoding. Second, it achieves efficient cross-subject generalization by sharing core expert networks while adapting only subject-specific routers. Third, it provides enhanced mechanistic insight, as the explicit routing reveals precisely how different modeled brain regions shape the semantic and spatial attributes of the reconstructed image. Extensive experiments validate MoRE-Brain's high reconstruction fidelity, with bottleneck analyses further demonstrating its effective utilization of fMRI signals, distinguishing genuine neural decoding from over-reliance on generative priors. Consequently, MoRE-Brain marks a substantial advance towards more generalizable and interpretable fMRI-based visual decoding. Code will be publicly available soon: https://github.com/yuxiangwei0808/MoRE-Brain.
4D Multimodal Co-attention Fusion Network with Latent Contrastive Alignment for Alzheimer's Diagnosis
Apr 23, 2025



Abstract:Multimodal neuroimaging provides complementary structural and functional insights into both human brain organization and disease-related dynamics. Recent studies demonstrate enhanced diagnostic sensitivity for Alzheimer's disease (AD) through synergistic integration of neuroimaging data (e.g., sMRI, fMRI) with behavioral cognitive scores tabular data biomarkers. However, the intrinsic heterogeneity across modalities (e.g., 4D spatiotemporal fMRI dynamics vs. 3D anatomical sMRI structure) presents critical challenges for discriminative feature fusion. To bridge this gap, we propose M2M-AlignNet: a geometry-aware multimodal co-attention network with latent alignment for early AD diagnosis using sMRI and fMRI. At the core of our approach is a multi-patch-to-multi-patch (M2M) contrastive loss function that quantifies and reduces representational discrepancies via geometry-weighted patch correspondence, explicitly aligning fMRI components across brain regions with their sMRI structural substrates without one-to-one constraints. Additionally, we propose a latent-as-query co-attention module to autonomously discover fusion patterns, circumventing modality prioritization biases while minimizing feature redundancy. We conduct extensive experiments to confirm the effectiveness of our method and highlight the correspondance between fMRI and sMRI as AD biomarkers.
A Privacy-Preserving Domain Adversarial Federated learning for multi-site brain functional connectivity analysis
Feb 03, 2025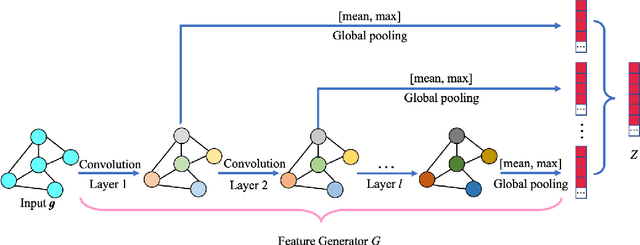
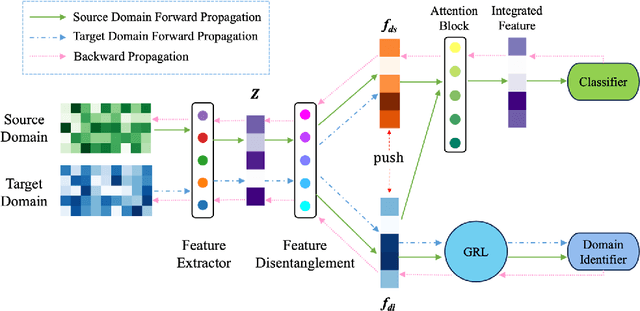
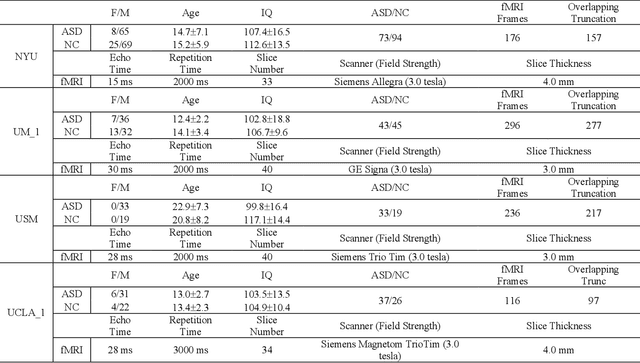
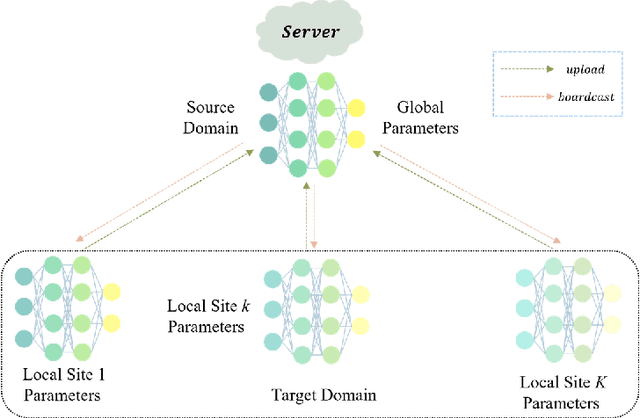
Abstract:Resting-state functional magnetic resonance imaging (rs-fMRI) and its derived functional connectivity networks (FCNs) have become critical for understanding neurological disorders. However, collaborative analyses and the generalizability of models still face significant challenges due to privacy regulations and the non-IID (non-independent and identically distributed) property of multiple data sources. To mitigate these difficulties, we propose Domain Adversarial Federated Learning (DAFed), a novel federated deep learning framework specifically designed for non-IID fMRI data analysis in multi-site settings. DAFed addresses these challenges through feature disentanglement, decomposing the latent feature space into domain-invariant and domain-specific components, to ensure robust global learning while preserving local data specificity. Furthermore, adversarial training facilitates effective knowledge transfer between labeled and unlabeled datasets, while a contrastive learning module enhances the global representation of domain-invariant features. We evaluated DAFed on the diagnosis of ASD and further validated its generalizability in the classification of AD, demonstrating its superior classification accuracy compared to state-of-the-art methods. Additionally, an enhanced Score-CAM module identifies key brain regions and functional connectivity significantly associated with ASD and MCI, respectively, uncovering shared neurobiological patterns across sites. These findings highlight the potential of DAFed to advance multi-site collaborative research in neuroimaging while protecting data confidentiality.
A Deep Spatio-Temporal Architecture for Dynamic Effective Connectivity Network Analysis Based on Dynamic Causal Discovery
Jan 31, 2025



Abstract:Dynamic effective connectivity networks (dECNs) reveal the changing directed brain activity and the dynamic causal influences among brain regions, which facilitate the identification of individual differences and enhance the understanding of human brain. Although the existing causal discovery methods have shown promising results in effective connectivity network analysis, they often overlook the dynamics of causality, in addition to the incorporation of spatio-temporal information in brain activity data. To address these issues, we propose a deep spatio-temporal fusion architecture, which employs a dynamic causal deep encoder to incorporate spatio-temporal information into dynamic causality modeling, and a dynamic causal deep decoder to verify the discovered causality. The effectiveness of the proposed method is first illustrated with simulated data. Then, experimental results from Philadelphia Neurodevelopmental Cohort (PNC) demonstrate the superiority of the proposed method in inferring dECNs, which reveal the dynamic evolution of directed flow between brain regions. The analysis shows the difference of dECNs between young adults and children. Specifically, the directed brain functional networks transit from fluctuating undifferentiated systems to more stable specialized networks as one grows. This observation provides further evidence on the modularization and adaptation of brain networks during development, leading to higher cognitive abilities observed in young adults.
Generative forecasting of brain activity enhances Alzheimer's classification and interpretation
Oct 30, 2024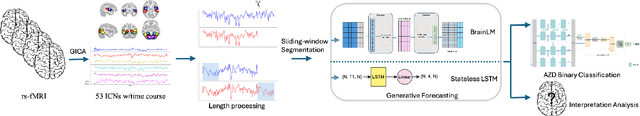
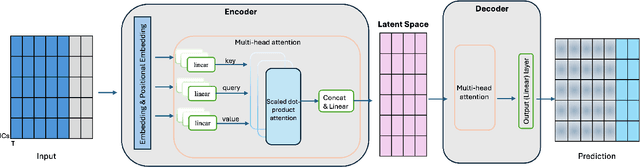
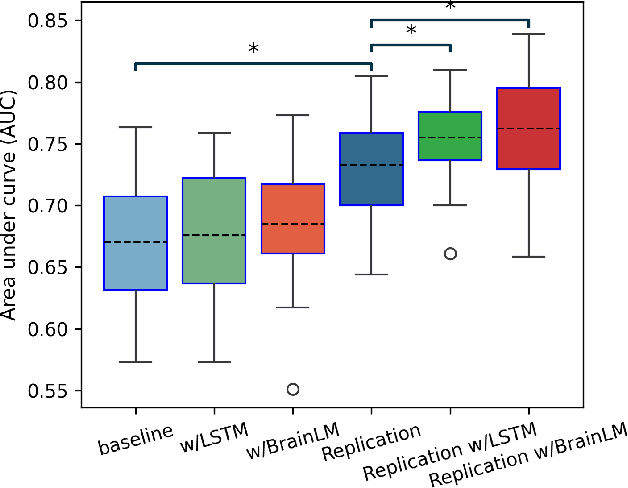
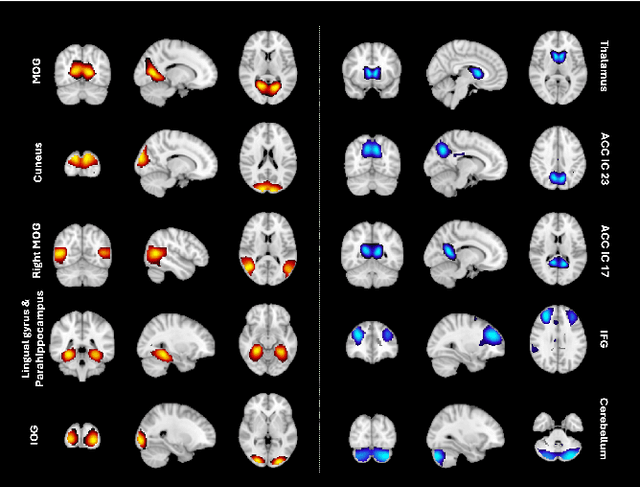
Abstract:Understanding the relationship between cognition and intrinsic brain activity through purely data-driven approaches remains a significant challenge in neuroscience. Resting-state functional magnetic resonance imaging (rs-fMRI) offers a non-invasive method to monitor regional neural activity, providing a rich and complex spatiotemporal data structure. Deep learning has shown promise in capturing these intricate representations. However, the limited availability of large datasets, especially for disease-specific groups such as Alzheimer's Disease (AD), constrains the generalizability of deep learning models. In this study, we focus on multivariate time series forecasting of independent component networks derived from rs-fMRI as a form of data augmentation, using both a conventional LSTM-based model and the novel Transformer-based BrainLM model. We assess their utility in AD classification, demonstrating how generative forecasting enhances classification performance. Post-hoc interpretation of BrainLM reveals class-specific brain network sensitivities associated with AD.
Integrated Brain Connectivity Analysis with fMRI, DTI, and sMRI Powered by Interpretable Graph Neural Networks
Aug 26, 2024Abstract:Multimodal neuroimaging modeling has becomes a widely used approach but confronts considerable challenges due to heterogeneity, which encompasses variability in data types, scales, and formats across modalities. This variability necessitates the deployment of advanced computational methods to integrate and interpret these diverse datasets within a cohesive analytical framework. In our research, we amalgamate functional magnetic resonance imaging, diffusion tensor imaging, and structural MRI into a cohesive framework. This integration capitalizes on the unique strengths of each modality and their inherent interconnections, aiming for a comprehensive understanding of the brain's connectivity and anatomical characteristics. Utilizing the Glasser atlas for parcellation, we integrate imaging derived features from various modalities: functional connectivity from fMRI, structural connectivity from DTI, and anatomical features from sMRI within consistent regions. Our approach incorporates a masking strategy to differentially weight neural connections, thereby facilitating a holistic amalgamation of multimodal imaging data. This technique enhances interpretability at connectivity level, transcending traditional analyses centered on singular regional attributes. The model is applied to the Human Connectome Project's Development study to elucidate the associations between multimodal imaging and cognitive functions throughout youth. The analysis demonstrates improved predictive accuracy and uncovers crucial anatomical features and essential neural connections, deepening our understanding of brain structure and function.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge