Julia M. Stephen
A Deep Spatio-Temporal Architecture for Dynamic Effective Connectivity Network Analysis Based on Dynamic Causal Discovery
Jan 31, 2025



Abstract:Dynamic effective connectivity networks (dECNs) reveal the changing directed brain activity and the dynamic causal influences among brain regions, which facilitate the identification of individual differences and enhance the understanding of human brain. Although the existing causal discovery methods have shown promising results in effective connectivity network analysis, they often overlook the dynamics of causality, in addition to the incorporation of spatio-temporal information in brain activity data. To address these issues, we propose a deep spatio-temporal fusion architecture, which employs a dynamic causal deep encoder to incorporate spatio-temporal information into dynamic causality modeling, and a dynamic causal deep decoder to verify the discovered causality. The effectiveness of the proposed method is first illustrated with simulated data. Then, experimental results from Philadelphia Neurodevelopmental Cohort (PNC) demonstrate the superiority of the proposed method in inferring dECNs, which reveal the dynamic evolution of directed flow between brain regions. The analysis shows the difference of dECNs between young adults and children. Specifically, the directed brain functional networks transit from fluctuating undifferentiated systems to more stable specialized networks as one grows. This observation provides further evidence on the modularization and adaptation of brain networks during development, leading to higher cognitive abilities observed in young adults.
A Demographic-Conditioned Variational Autoencoder for fMRI Distribution Sampling and Removal of Confounds
May 13, 2024



Abstract:Objective: fMRI and derived measures such as functional connectivity (FC) have been used to predict brain age, general fluid intelligence, psychiatric disease status, and preclinical neurodegenerative disease. However, it is not always clear that all demographic confounds, such as age, sex, and race, have been removed from fMRI data. Additionally, many fMRI datasets are restricted to authorized researchers, making dissemination of these valuable data sources challenging. Methods: We create a variational autoencoder (VAE)-based model, DemoVAE, to decorrelate fMRI features from demographics and generate high-quality synthetic fMRI data based on user-supplied demographics. We train and validate our model using two large, widely used datasets, the Philadelphia Neurodevelopmental Cohort (PNC) and Bipolar and Schizophrenia Network for Intermediate Phenotypes (BSNIP). Results: We find that DemoVAE recapitulates group differences in fMRI data while capturing the full breadth of individual variations. Significantly, we also find that most clinical and computerized battery fields that are correlated with fMRI data are not correlated with DemoVAE latents. An exception are several fields related to schizophrenia medication and symptom severity. Conclusion: Our model generates fMRI data that captures the full distribution of FC better than traditional VAE or GAN models. We also find that most prediction using fMRI data is dependent on correlation with, and prediction of, demographics. Significance: Our DemoVAE model allows for generation of high quality synthetic data conditioned on subject demographics as well as the removal of the confounding effects of demographics. We identify that FC-based prediction tasks are highly influenced by demographic confounds.
Distance Correlation Based Brain Functional Connectivity Estimation and Non-Convex Multi-Task Learning for Developmental fMRI Studies
Sep 30, 2020



Abstract:Resting-state functional magnetic resonance imaging (rs-fMRI)-derived functional connectivity patterns have been extensively utilized to delineate global functional organization of the human brain in health, development, and neuropsychiatric disorders. In this paper, we investigate how functional connectivity in males and females differs in an age prediction framework. We first estimate functional connectivity between regions-of-interest (ROIs) using distance correlation instead of Pearson's correlation. Distance correlation, as a multivariate statistical method, explores spatial relations of voxel-wise time courses within individual ROIs and measures both linear and nonlinear dependence, capturing more complex information of between-ROI interactions. Then, a novel non-convex multi-task learning (NC-MTL) model is proposed to study age-related gender differences in functional connectivity, where age prediction for each gender group is viewed as one task. Specifically, in the proposed NC-MTL model, we introduce a composite regularizer with a combination of non-convex $\ell_{2,1-2}$ and $\ell_{1-2}$ regularization terms for selecting both common and task-specific features. Finally, we validate the proposed NC-MTL model along with distance correlation based functional connectivity on rs-fMRI of the Philadelphia Neurodevelopmental Cohort for predicting ages of both genders. The experimental results demonstrate that the proposed NC-MTL model outperforms other competing MTL models in age prediction, as well as characterizing developmental gender differences in functional connectivity patterns.
Causal inference of brain connectivity from fMRI with $ψ$-Learning Incorporated Linear non-Gaussian Acyclic Model ($ψ$-LiNGAM)
Jun 16, 2020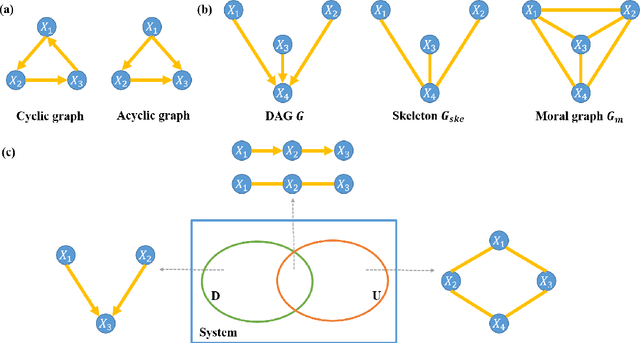
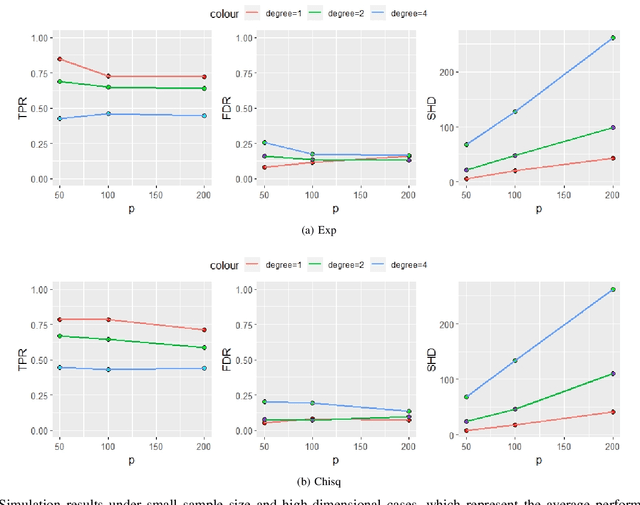
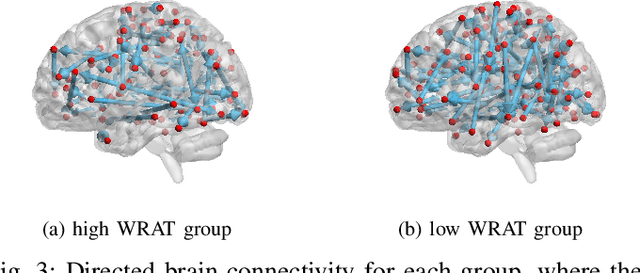
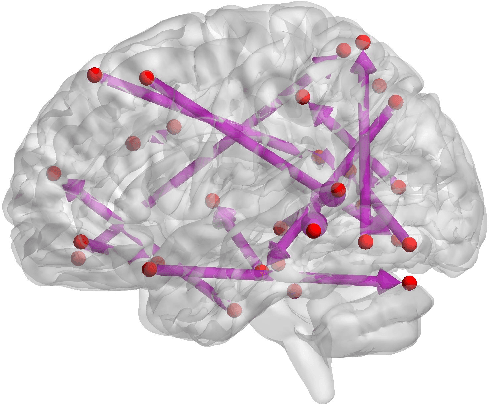
Abstract:Functional connectivity (FC) has become a primary means of understanding brain functions by identifying brain network interactions and, ultimately, how those interactions produce cognitions. A popular definition of FC is by statistical associations between measured brain regions. However, this could be problematic since the associations can only provide spatial connections but not causal interactions among regions of interests. Hence, it is necessary to study their causal relationship. Directed acyclic graph (DAG) models have been applied in recent FC studies but often encountered problems such as limited sample sizes and large number of variables (namely high-dimensional problems), which lead to both computational difficulty and convergence issues. As a result, the use of DAG models is problematic, where the identification of DAG models in general is nondeterministic polynomial time hard (NP-hard). To this end, we propose a $\psi$-learning incorporated linear non-Gaussian acyclic model ($\psi$-LiNGAM). We use the association model ($\psi$-learning) to facilitate causal inferences and the model works well especially for high-dimensional cases. Our simulation results demonstrate that the proposed method is more robust and accurate than several existing ones in detecting graph structure and direction. We then applied it to the resting state fMRI (rsfMRI) data obtained from the publicly available Philadelphia Neurodevelopmental Cohort (PNC) to study the cognitive variance, which includes 855 individuals aged 8-22 years. Therein, we have identified three types of hub structure: the in-hub, out-hub and sum-hub, which correspond to the centers of receiving, sending and relaying information, respectively. We also detected 16 most important pairs of causal flows. Several of the results have been verified to be biologically significant.
A Bayesian incorporated linear non-Gaussian acyclic model for multiple directed graph estimation to study brain emotion circuit development in adolescence
Jun 16, 2020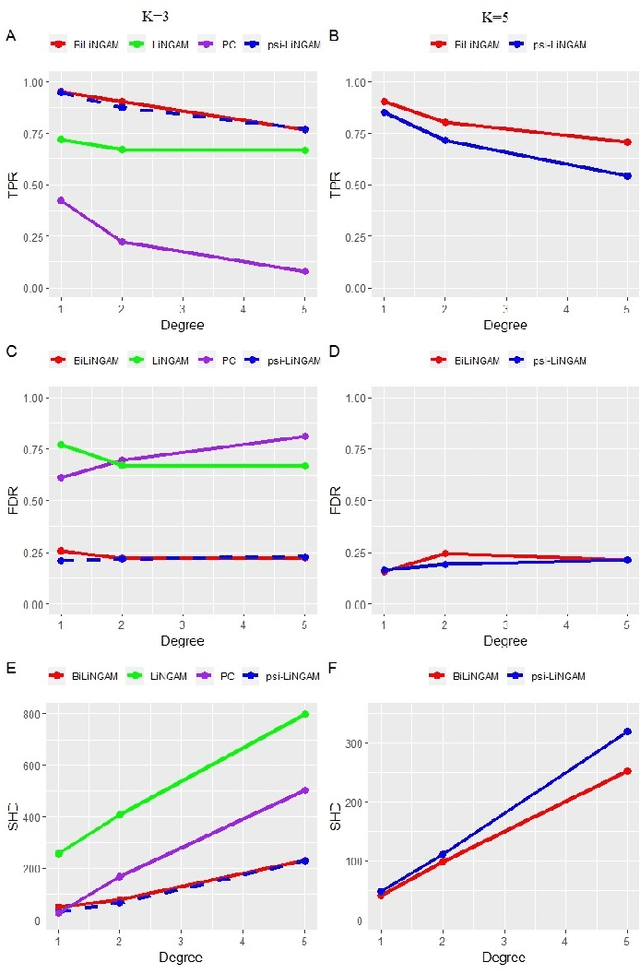
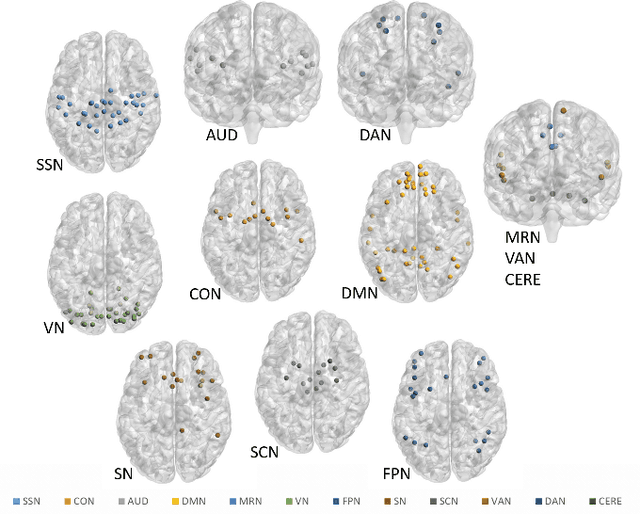
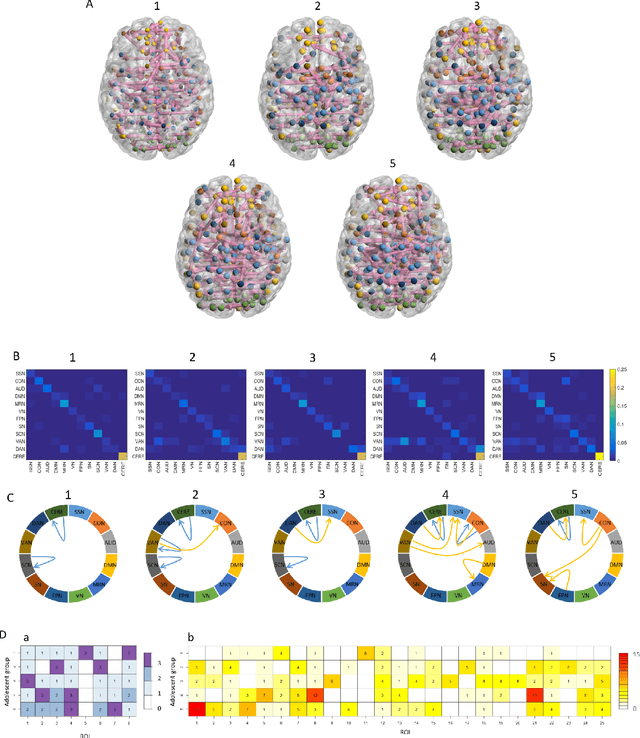
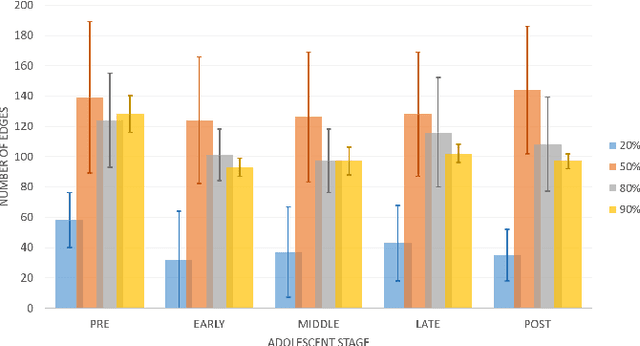
Abstract:Emotion perception is essential to affective and cognitive development which involves distributed brain circuits. The ability of emotion identification begins in infancy and continues to develop throughout childhood and adolescence. Understanding the development of brain's emotion circuitry may help us explain the emotional changes observed during adolescence. Our previous study delineated the trajectory of brain functional connectivity (FC) from late childhood to early adulthood during emotion identification tasks. In this work, we endeavour to deepen our understanding from association to causation. We proposed a Bayesian incorporated linear non-Gaussian acyclic model (BiLiNGAM), which incorporated our previous association model into the prior estimation pipeline. In particular, it can jointly estimate multiple directed acyclic graphs (DAGs) for multiple age groups at different developmental stages. Simulation results indicated more stable and accurate performance over various settings, especially when the sample size was small (high-dimensional cases). We then applied to the analysis of real data from the Philadelphia Neurodevelopmental Cohort (PNC). This included 855 individuals aged 8-22 years who were divided into five different adolescent stages. Our network analysis revealed the development of emotion-related intra- and inter- modular connectivity and pinpointed several emotion-related hubs. We further categorized the hubs into two types: in-hubs and out-hubs, as the center of receiving and distributing information. Several unique developmental hub structures and group-specific patterns were also discovered. Our findings help provide a causal understanding of emotion development in the human brain.
Interpretable multimodal fusion networks reveal mechanisms of brain cognition
Jun 16, 2020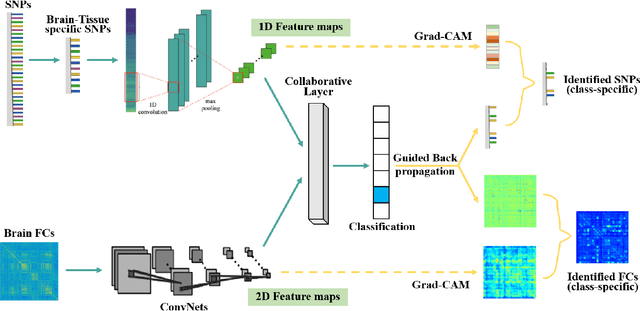
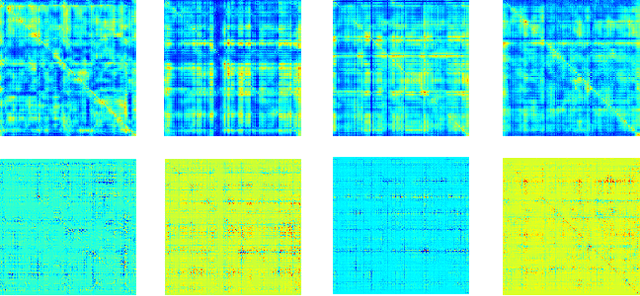
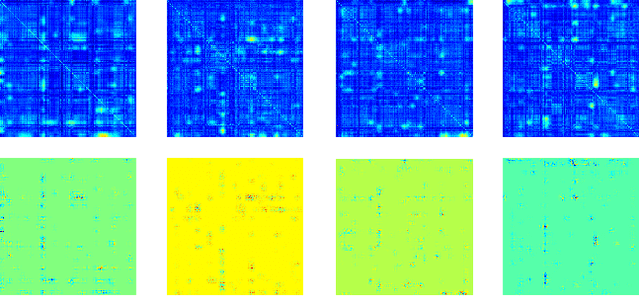
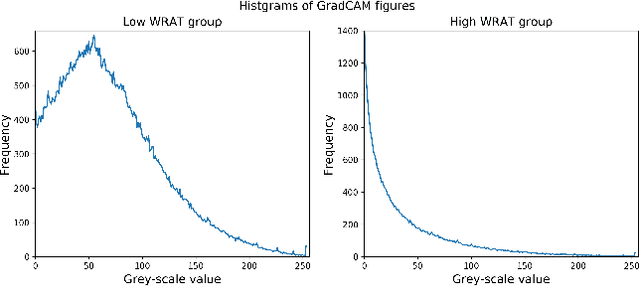
Abstract:Multimodal fusion benefits disease diagnosis by providing a more comprehensive perspective. Developing algorithms is challenging due to data heterogeneity and the complex within- and between-modality associations. Deep-network-based data-fusion models have been developed to capture the complex associations and the performance in diagnosis has been improved accordingly. Moving beyond diagnosis prediction, evaluation of disease mechanisms is critically important for biomedical research. Deep-network-based data-fusion models, however, are difficult to interpret, bringing about difficulties for studying biological mechanisms. In this work, we develop an interpretable multimodal fusion model, namely gCAM-CCL, which can perform automated diagnosis and result interpretation simultaneously. The gCAM-CCL model can generate interpretable activation maps, which quantify pixel-level contributions of the input features. This is achieved by combining intermediate feature maps using gradient-based weights. Moreover, the estimated activation maps are class-specific, and the captured cross-data associations are interest/label related, which further facilitates class-specific analysis and biological mechanism analysis. We validate the gCAM-CCL model on a brain imaging-genetic study, and show gCAM-CCL's performed well for both classification and mechanism analysis. Mechanism analysis suggests that during task-fMRI scans, several object recognition related regions of interests (ROIs) are first activated and then several downstream encoding ROIs get involved. Results also suggest that the higher cognition performing group may have stronger neurotransmission signaling while the lower cognition performing group may have problem in brain/neuron development, resulting from genetic variations.
Causality based Feature Fusion for Brain Neuro-Developmental Analysis
Jan 22, 2020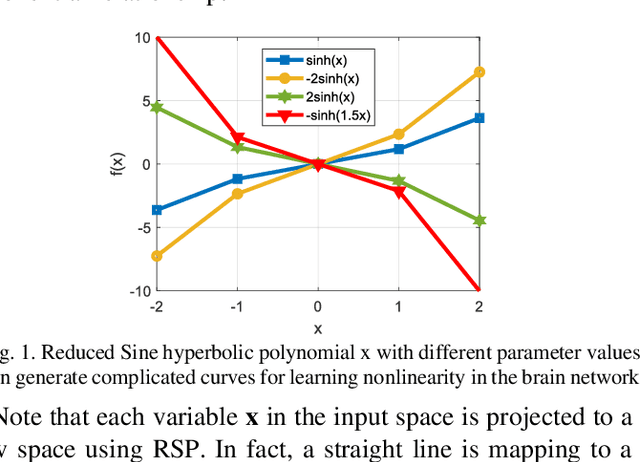
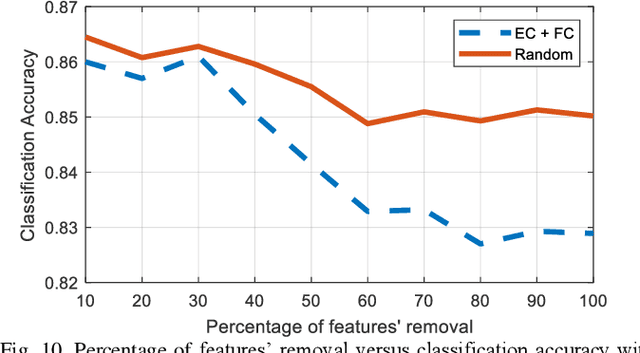
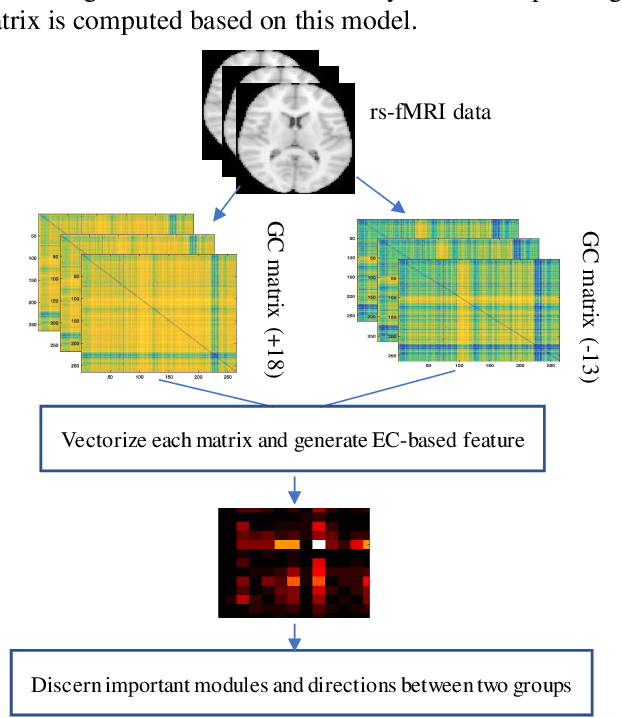
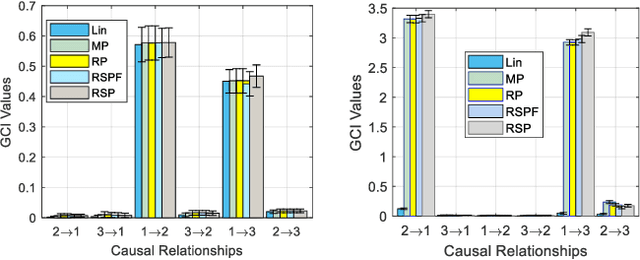
Abstract:Human brain development is a complex and dynamic process that is affected by several factors such as genetics, sex hormones, and environmental changes. A number of recent studies on brain development have examined functional connectivity (FC) defined by the temporal correlation between time series of different brain regions. We propose to add the directional flow of information during brain maturation. To do so, we extract effective connectivity (EC) through Granger causality (GC) for two different groups of subjects, i.e., children and young adults. The motivation is that the inclusion of causal interaction may further discriminate brain connections between two age groups and help to discover new connections between brain regions. The contributions of this study are threefold. First, there has been a lack of attention to EC-based feature extraction in the context of brain development. To this end, we propose a new kernel-based GC (KGC) method to learn nonlinearity of complex brain network, where a reduced Sine hyperbolic polynomial (RSP) neural network was used as our proposed learner. Second, we used causality values as the weight for the directional connectivity between brain regions. Our findings indicated that the strength of connections was significantly higher in young adults relative to children. In addition, our new EC-based feature outperformed FC-based analysis from Philadelphia neurocohort (PNC) study with better discrimination of the different age groups. Moreover, the fusion of these two sets of features (FC + EC) improved brain age prediction accuracy by more than 4%, indicating that they should be used together for brain development studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge