Ziyu Zhou
SEDformer: Event-Synchronous Spiking Transformers for Irregular Telemetry Time Series Forecasting
Feb 03, 2026Abstract:Telemetry streams from large-scale Internet-connected systems (e.g., IoT deployments and online platforms) naturally form an irregular multivariate time series (IMTS) whose accurate forecasting is operationally vital. A closer examination reveals a defining Sparsity-Event Duality (SED) property of IMTS, i.e., long stretches with sparse or no observations are punctuated by short, dense bursts where most semantic events (observations) occur. However, existing Graph- and Transformer-based forecasters ignore SED: pre-alignment to uniform grids with heavy padding violates sparsity by inflating sequences and forcing computation at non-informative steps, while relational recasting weakens event semantics by disrupting local temporal continuity. These limitations motivate a more faithful and natural modeling paradigm for IMTS that aligns with its SED property. We find that Spiking Neural Networks meet this requirement, as they communicate via sparse binary spikes and update in an event-driven manner, aligning naturally with the SED nature of IMTS. Therefore, we present SEDformer, an SED-enhanced Spiking Transformer for telemetry IMTS forecasting that couples: (1) a SED-based Spike Encoder converts raw observations into event synchronous spikes using an Event-Aligned LIF neuron, (2) an Event-Preserving Temporal Downsampling module compresses long gaps while retaining salient firings and (3) a stack of SED-based Spike Transformer blocks enable intra-series dependency modeling with a membrane-based linear attention driven by EA-LIF spiking features. Experiments on public telemetry IMTS datasets show that SEDformer attains state-of-the-art forecasting accuracy while reducing energy and memory usage, providing a natural and efficient path for modeling IMTS.
WordCraft: Scaffolding the Keyword Method for L2 Vocabulary Learning with Multimodal LLMs
Jan 31, 2026Abstract:Applying the keyword method for vocabulary memorization remains a significant challenge for L1 Chinese-L2 English learners. They frequently struggle to generate phonologically appropriate keywords, construct coherent associations, and create vivid mental imagery to aid long-term retention. Existing approaches, including fully automated keyword generation and outcome-oriented mnemonic aids, either compromise learner engagement or lack adequate process-oriented guidance. To address these limitations, we conducted a formative study with L1 Chinese-L2 English learners and educators (N=18), which revealed key difficulties and requirements in applying the keyword method to vocabulary learning. Building on these insights, we introduce WordCraft, a learner-centered interactive tool powered by Multimodal Large Language Models (MLLMs). WordCraft scaffolds the keyword method by guiding learners through keyword selection, association construction, and image formation, thereby enhancing the effectiveness of vocabulary memorization. Two user studies demonstrate that WordCraft not only preserves the generation effect but also achieves high levels of effectiveness and usability.
Lamps: Learning Anatomy from Multiple Perspectives via Self-supervision in Chest Radiographs
Jan 02, 2026Abstract:Foundation models have been successful in natural language processing and computer vision because they are capable of capturing the underlying structures (foundation) of natural languages. However, in medical imaging, the key foundation lies in human anatomy, as these images directly represent the internal structures of the body, reflecting the consistency, coherence, and hierarchy of human anatomy. Yet, existing self-supervised learning (SSL) methods often overlook these perspectives, limiting their ability to effectively learn anatomical features. To overcome the limitation, we built Lamps (learning anatomy from multiple perspectives via self-supervision) pre-trained on large-scale chest radiographs by harmoniously utilizing the consistency, coherence, and hierarchy of human anatomy as the supervision signal. Extensive experiments across 10 datasets evaluated through fine-tuning and emergent property analysis demonstrate Lamps' superior robustness, transferability, and clinical potential when compared to 10 baseline models. By learning from multiple perspectives, Lamps presents a unique opportunity for foundation models to develop meaningful, robust representations that are aligned with the structure of human anatomy.
Learning Anatomy from Multiple Perspectives via Self-supervision in Chest Radiographs
Dec 28, 2025Abstract:Foundation models have been successful in natural language processing and computer vision because they are capable of capturing the underlying structures (foundation) of natural languages. However, in medical imaging, the key foundation lies in human anatomy, as these images directly represent the internal structures of the body, reflecting the consistency, coherence, and hierarchy of human anatomy. Yet, existing self-supervised learning (SSL) methods often overlook these perspectives, limiting their ability to effectively learn anatomical features. To overcome the limitation, we built Lamps (learning anatomy from multiple perspectives via self-supervision) pre-trained on large-scale chest radiographs by harmoniously utilizing the consistency, coherence, and hierarchy of human anatomy as the supervision signal. Extensive experiments across 10 datasets evaluated through fine-tuning and emergent property analysis demonstrate Lamps' superior robustness, transferability, and clinical potential when compared to 10 baseline models. By learning from multiple perspectives, Lamps presents a unique opportunity for foundation models to develop meaningful, robust representations that are aligned with the structure of human anatomy.
Multi-Order Wavelet Derivative Transform for Deep Time Series Forecasting
May 17, 2025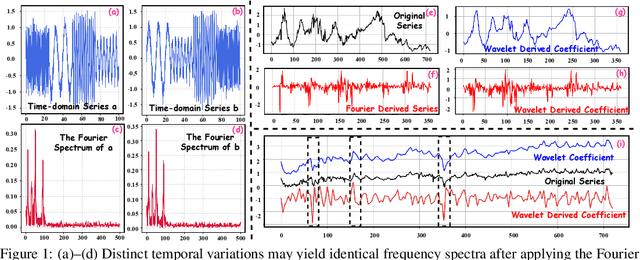
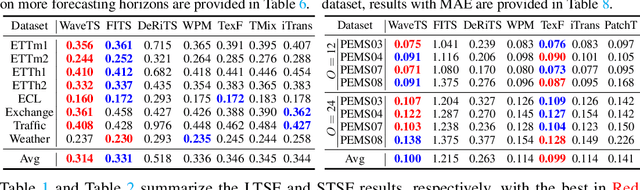
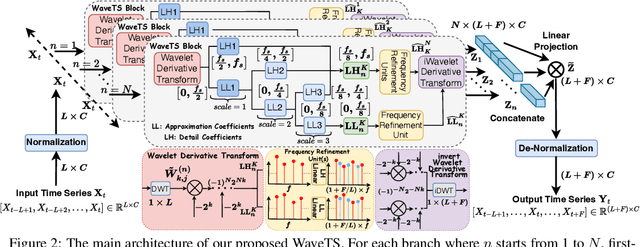
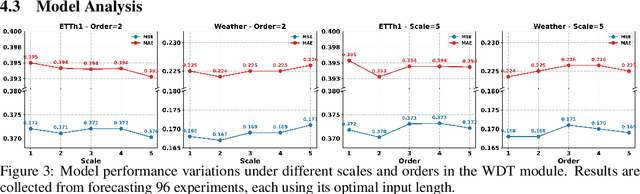
Abstract:In deep time series forecasting, the Fourier Transform (FT) is extensively employed for frequency representation learning. However, it often struggles in capturing multi-scale, time-sensitive patterns. Although the Wavelet Transform (WT) can capture these patterns through frequency decomposition, its coefficients are insensitive to change points in time series, leading to suboptimal modeling. To mitigate these limitations, we introduce the multi-order Wavelet Derivative Transform (WDT) grounded in the WT, enabling the extraction of time-aware patterns spanning both the overall trend and subtle fluctuations. Compared with the standard FT and WT, which model the raw series, the WDT operates on the derivative of the series, selectively magnifying rate-of-change cues and exposing abrupt regime shifts that are particularly informative for time series modeling. Practically, we embed the WDT into a multi-branch framework named WaveTS, which decomposes the input series into multi-scale time-frequency coefficients, refines them via linear layers, and reconstructs them into the time domain via the inverse WDT. Extensive experiments on ten benchmark datasets demonstrate that WaveTS achieves state-of-the-art forecasting accuracy while retaining high computational efficiency.
On the Interplay of Human-AI Alignment,Fairness, and Performance Trade-offs in Medical Imaging
May 15, 2025Abstract:Deep neural networks excel in medical imaging but remain prone to biases, leading to fairness gaps across demographic groups. We provide the first systematic exploration of Human-AI alignment and fairness in this domain. Our results show that incorporating human insights consistently reduces fairness gaps and enhances out-of-domain generalization, though excessive alignment can introduce performance trade-offs, emphasizing the need for calibrated strategies. These findings highlight Human-AI alignment as a promising approach for developing fair, robust, and generalizable medical AI systems, striking a balance between expert guidance and automated efficiency. Our code is available at https://github.com/Roypic/Aligner.
Evaluating the Effectiveness of Black-Box Prompt Optimization as the Scale of LLMs Continues to Grow
May 13, 2025Abstract:Black-Box prompt optimization methods have emerged as a promising strategy for refining input prompts to better align large language models (LLMs), thereby enhancing their task performance. Although these methods have demonstrated encouraging results, most studies and experiments have primarily focused on smaller-scale models (e.g., 7B, 14B) or earlier versions (e.g., GPT-3.5) of LLMs. As the scale of LLMs continues to increase, such as with DeepSeek V3 (671B), it remains an open question whether these black-box optimization techniques will continue to yield significant performance improvements for models of such scale. In response to this, we select three well-known black-box optimization methods and evaluate them on large-scale LLMs (DeepSeek V3 and Gemini 2.0 Flash) across four NLU and NLG datasets. The results show that these black-box prompt optimization methods offer only limited improvements on these large-scale LLMs. Furthermore, we hypothesize that the scale of the model is the primary factor contributing to the limited benefits observed. To explore this hypothesis, we conducted experiments on LLMs of varying sizes (Qwen 2.5 series, ranging from 7B to 72B) and observed an inverse scaling law, wherein the effectiveness of black-box optimization methods diminished as the model size increased.
Neural Approaches to SAT Solving: Design Choices and Interpretability
Apr 01, 2025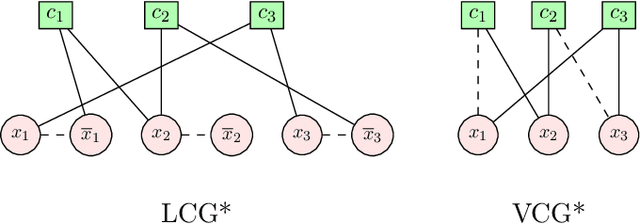
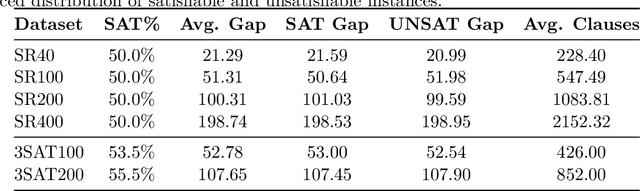
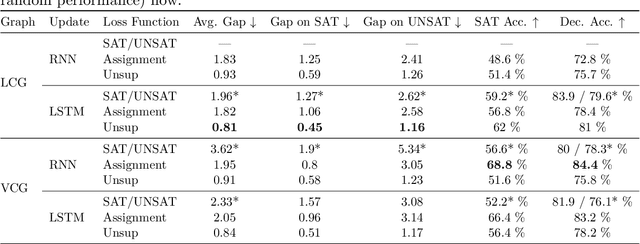
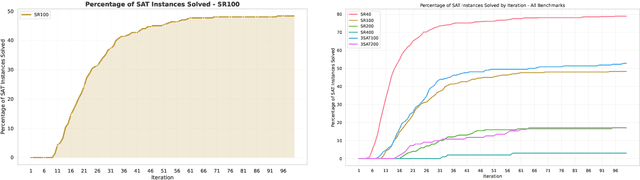
Abstract:In this contribution, we provide a comprehensive evaluation of graph neural networks applied to Boolean satisfiability problems, accompanied by an intuitive explanation of the mechanisms enabling the model to generalize to different instances. We introduce several training improvements, particularly a novel closest assignment supervision method that dynamically adapts to the model's current state, significantly enhancing performance on problems with larger solution spaces. Our experiments demonstrate the suitability of variable-clause graph representations with recurrent neural network updates, which achieve good accuracy on SAT assignment prediction while reducing computational demands. We extend the base graph neural network into a diffusion model that facilitates incremental sampling and can be effectively combined with classical techniques like unit propagation. Through analysis of embedding space patterns and optimization trajectories, we show how these networks implicitly perform a process very similar to continuous relaxations of MaxSAT, offering an interpretable view of their reasoning process. This understanding guides our design choices and explains the ability of recurrent architectures to scale effectively at inference time beyond their training distribution, which we demonstrate with test-time scaling experiments.
WorkTeam: Constructing Workflows from Natural Language with Multi-Agents
Mar 28, 2025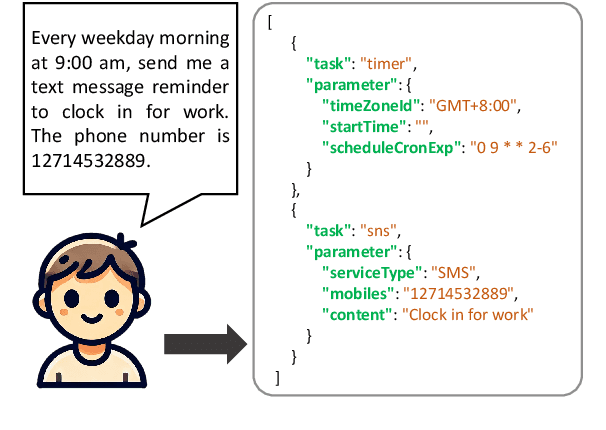
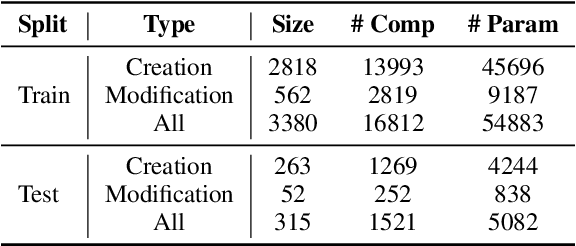
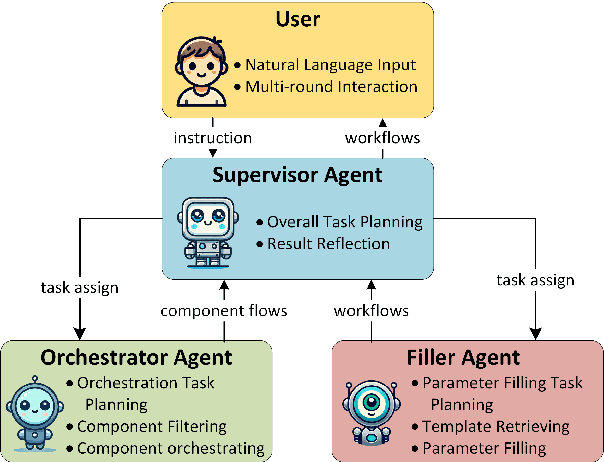
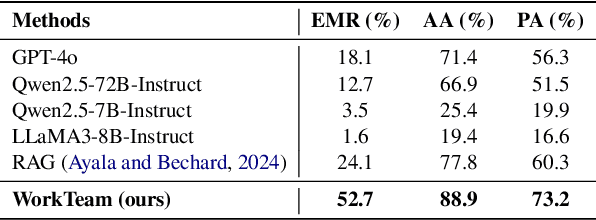
Abstract:Workflows play a crucial role in enhancing enterprise efficiency by orchestrating complex processes with multiple tools or components. However, hand-crafted workflow construction requires expert knowledge, presenting significant technical barriers. Recent advancements in Large Language Models (LLMs) have improved the generation of workflows from natural language instructions (aka NL2Workflow), yet existing single LLM agent-based methods face performance degradation on complex tasks due to the need for specialized knowledge and the strain of task-switching. To tackle these challenges, we propose WorkTeam, a multi-agent NL2Workflow framework comprising a supervisor, orchestrator, and filler agent, each with distinct roles that collaboratively enhance the conversion process. As there are currently no publicly available NL2Workflow benchmarks, we also introduce the HW-NL2Workflow dataset, which includes 3,695 real-world business samples for training and evaluation. Experimental results show that our approach significantly increases the success rate of workflow construction, providing a novel and effective solution for enterprise NL2Workflow services.
SChanger: Change Detection from a Semantic Change and Spatial Consistency Perspective
Mar 26, 2025Abstract:Change detection is a key task in Earth observation applications. Recently, deep learning methods have demonstrated strong performance and widespread application. However, change detection faces data scarcity due to the labor-intensive process of accurately aligning remote sensing images of the same area, which limits the performance of deep learning algorithms. To address the data scarcity issue, we develop a fine-tuning strategy called the Semantic Change Network (SCN). We initially pre-train the model on single-temporal supervised tasks to acquire prior knowledge of instance feature extraction. The model then employs a shared-weight Siamese architecture and extended Temporal Fusion Module (TFM) to preserve this prior knowledge and is fine-tuned on change detection tasks. The learned semantics for identifying all instances is changed to focus on identifying only the changes. Meanwhile, we observe that the locations of changes between the two images are spatially identical, a concept we refer to as spatial consistency. We introduce this inductive bias through an attention map that is generated by large-kernel convolutions and applied to the features from both time points. This enhances the modeling of multi-scale changes and helps capture underlying relationships in change detection semantics. We develop a binary change detection model utilizing these two strategies. The model is validated against state-of-the-art methods on six datasets, surpassing all benchmark methods and achieving F1 scores of 92.87%, 86.43%, 68.95%, 97.62%, 84.58%, and 93.20% on the LEVIR-CD, LEVIR-CD+, S2Looking, CDD, SYSU-CD, and WHU-CD datasets, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge