Wenxing Hu
SAFE-QAQ: End-to-End Slow-Thinking Audio-Text Fraud Detection via Reinforcement Learning
Jan 04, 2026Abstract:Existing fraud detection methods predominantly rely on transcribed text, suffering from ASR errors and missing crucial acoustic cues like vocal tone and environmental context. This limits their effectiveness against complex deceptive strategies. To address these challenges, we first propose \textbf{SAFE-QAQ}, an end-to-end comprehensive framework for audio-based slow-thinking fraud detection. First, the SAFE-QAQ framework eliminates the impact of transcription errors on detection performance. Secondly, we propose rule-based slow-thinking reward mechanisms that systematically guide the system to identify fraud-indicative patterns by accurately capturing fine-grained audio details, through hierarchical reasoning processes. Besides, our framework introduces a dynamic risk assessment framework during live calls, enabling early detection and prevention of fraud. Experiments on the TeleAntiFraud-Bench demonstrate that SAFE-QAQ achieves dramatic improvements over existing methods in multiple key dimensions, including accuracy, inference efficiency, and real-time processing capabilities. Currently deployed and analyzing over 70,000 calls daily, SAFE-QAQ effectively automates complex fraud detection, reducing human workload and financial losses. Code: https://anonymous.4open.science/r/SAFE-QAQ.
TRR360D: A dataset for 360 degree rotated rectangular box table detection
Mar 08, 2023
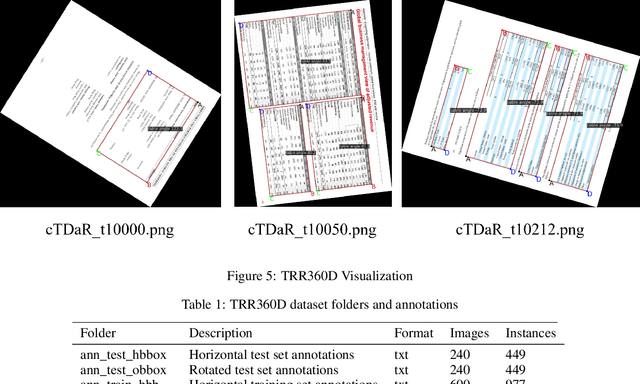


Abstract:To address the problem of scarcity and high annotation costs of rotated image table detection datasets, this paper proposes a method for building a rotated image table detection dataset. Based on the ICDAR2019MTD modern table detection dataset, we refer to the annotation format of the DOTA dataset to create the TRR360D rotated table detection dataset. The training set contains 600 rotated images and 977 annotated instances, and the test set contains 240 rotated images and 499 annotated instances. The AP50(T<90) evaluation metric is defined, and this dataset is available for future researchers to study rotated table detection algorithms and promote the development of table detection technology. The TRR360D rotated table detection dataset was created by constraining the starting point and annotation direction, and is publicly available at https://github.com/vansin/TRR360D.
Ensemble manifold based regularized multi-modal graph convolutional network for cognitive ability prediction
Jan 20, 2021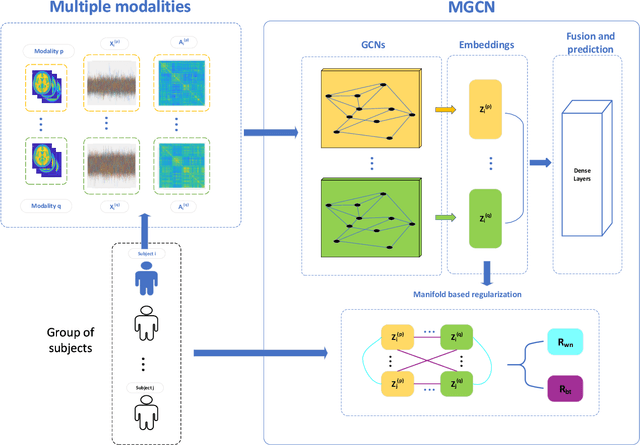
Abstract:Objective: Multi-modal functional magnetic resonance imaging (fMRI) can be used to make predictions about individual behavioral and cognitive traits based on brain connectivity networks. Methods: To take advantage of complementary information from multi-modal fMRI, we propose an interpretable multi-modal graph convolutional network (MGCN) model, incorporating the fMRI time series and the functional connectivity (FC) between each pair of brain regions. Specifically, our model learns a graph embedding from individual brain networks derived from multi-modal data. A manifold-based regularization term is then enforced to consider the relationships of subjects both within and between modalities. Furthermore, we propose the gradient-weighted regression activation mapping (Grad-RAM) and the edge mask learning to interpret the model, which is used to identify significant cognition-related biomarkers. Results: We validate our MGCN model on the Philadelphia Neurodevelopmental Cohort to predict individual wide range achievement test (WRAT) score. Our model obtains superior predictive performance over GCN with a single modality and other competing approaches. The identified biomarkers are cross-validated from different approaches. Conclusion and Significance: This paper develops a new interpretable graph deep learning framework for cognitive ability prediction, with the potential to overcome the limitations of several current data-fusion models. The results demonstrate the power of MGCN in analyzing multi-modal fMRI and discovering significant biomarkers for human brain studies.
Causal inference of brain connectivity from fMRI with $ψ$-Learning Incorporated Linear non-Gaussian Acyclic Model ($ψ$-LiNGAM)
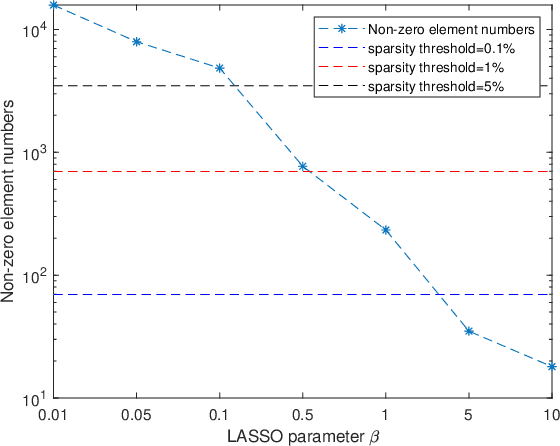
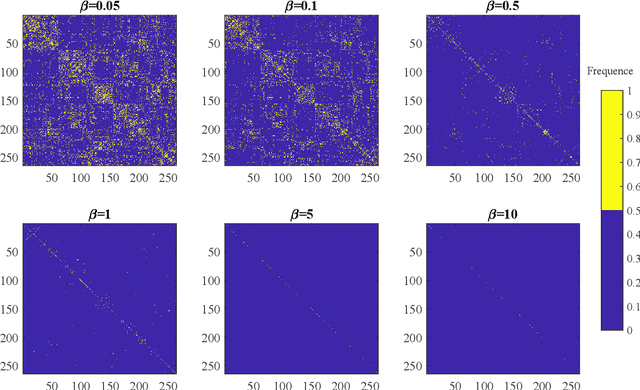
Jun 16, 2020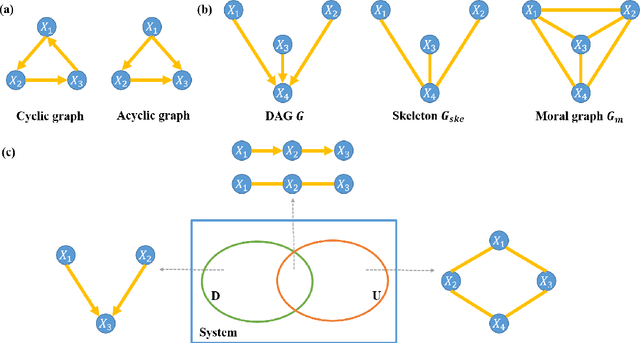
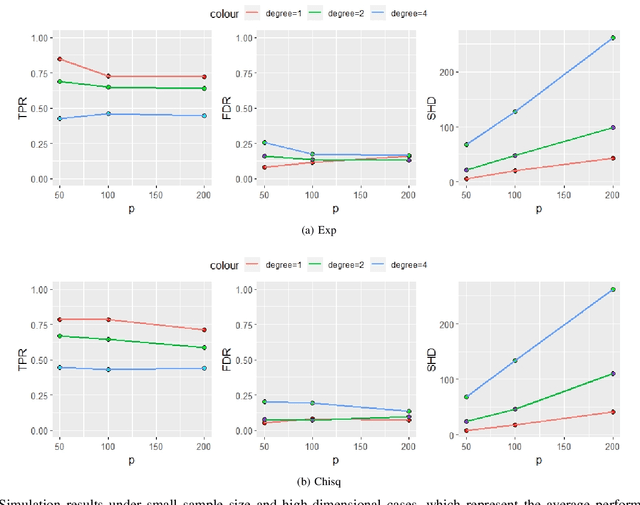
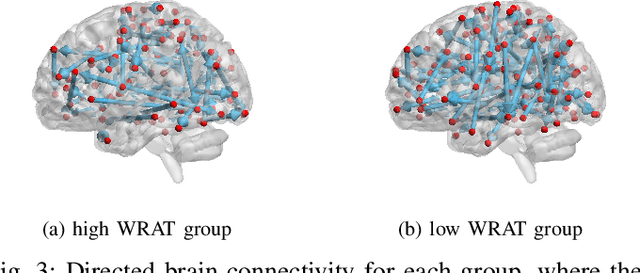
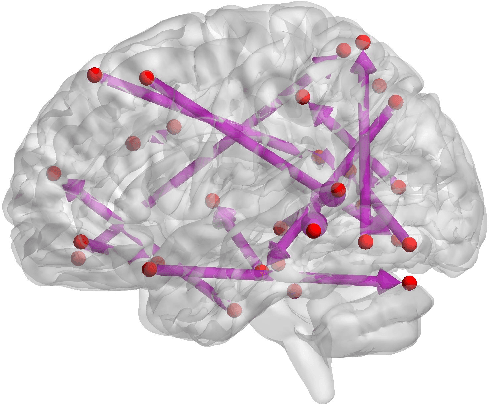
Abstract:Functional connectivity (FC) has become a primary means of understanding brain functions by identifying brain network interactions and, ultimately, how those interactions produce cognitions. A popular definition of FC is by statistical associations between measured brain regions. However, this could be problematic since the associations can only provide spatial connections but not causal interactions among regions of interests. Hence, it is necessary to study their causal relationship. Directed acyclic graph (DAG) models have been applied in recent FC studies but often encountered problems such as limited sample sizes and large number of variables (namely high-dimensional problems), which lead to both computational difficulty and convergence issues. As a result, the use of DAG models is problematic, where the identification of DAG models in general is nondeterministic polynomial time hard (NP-hard). To this end, we propose a $\psi$-learning incorporated linear non-Gaussian acyclic model ($\psi$-LiNGAM). We use the association model ($\psi$-learning) to facilitate causal inferences and the model works well especially for high-dimensional cases. Our simulation results demonstrate that the proposed method is more robust and accurate than several existing ones in detecting graph structure and direction. We then applied it to the resting state fMRI (rsfMRI) data obtained from the publicly available Philadelphia Neurodevelopmental Cohort (PNC) to study the cognitive variance, which includes 855 individuals aged 8-22 years. Therein, we have identified three types of hub structure: the in-hub, out-hub and sum-hub, which correspond to the centers of receiving, sending and relaying information, respectively. We also detected 16 most important pairs of causal flows. Several of the results have been verified to be biologically significant.
Interpretable multimodal fusion networks reveal mechanisms of brain cognition
Jun 16, 2020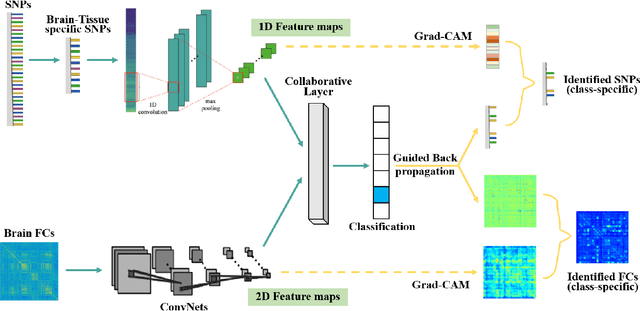
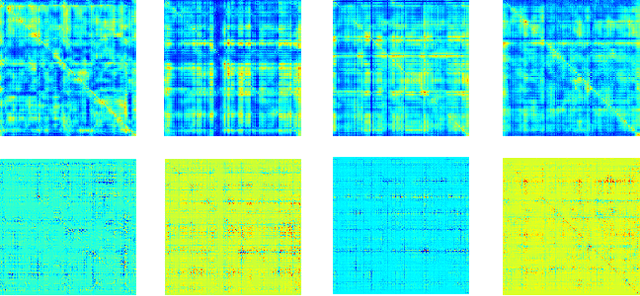
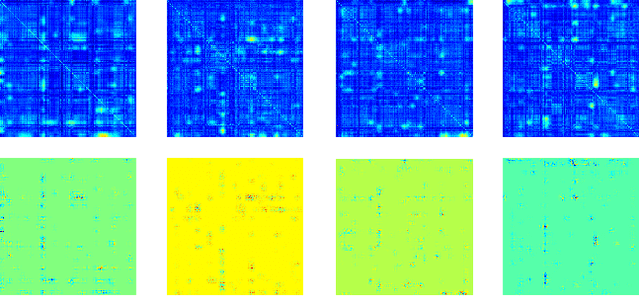
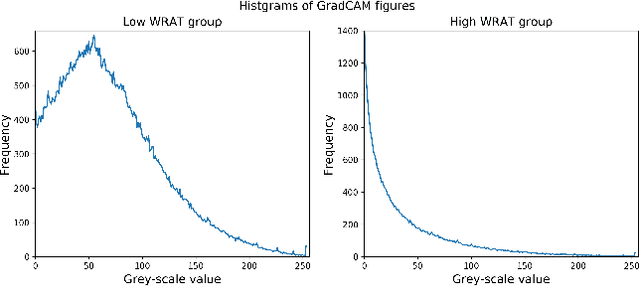
Abstract:Multimodal fusion benefits disease diagnosis by providing a more comprehensive perspective. Developing algorithms is challenging due to data heterogeneity and the complex within- and between-modality associations. Deep-network-based data-fusion models have been developed to capture the complex associations and the performance in diagnosis has been improved accordingly. Moving beyond diagnosis prediction, evaluation of disease mechanisms is critically important for biomedical research. Deep-network-based data-fusion models, however, are difficult to interpret, bringing about difficulties for studying biological mechanisms. In this work, we develop an interpretable multimodal fusion model, namely gCAM-CCL, which can perform automated diagnosis and result interpretation simultaneously. The gCAM-CCL model can generate interpretable activation maps, which quantify pixel-level contributions of the input features. This is achieved by combining intermediate feature maps using gradient-based weights. Moreover, the estimated activation maps are class-specific, and the captured cross-data associations are interest/label related, which further facilitates class-specific analysis and biological mechanism analysis. We validate the gCAM-CCL model on a brain imaging-genetic study, and show gCAM-CCL's performed well for both classification and mechanism analysis. Mechanism analysis suggests that during task-fMRI scans, several object recognition related regions of interests (ROIs) are first activated and then several downstream encoding ROIs get involved. Results also suggest that the higher cognition performing group may have stronger neurotransmission signaling while the lower cognition performing group may have problem in brain/neuron development, resulting from genetic variations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge