Causal inference of brain connectivity from fMRI with $ψ$-Learning Incorporated Linear non-Gaussian Acyclic Model ($ψ$-LiNGAM)
Paper and Code
Jun 16, 2020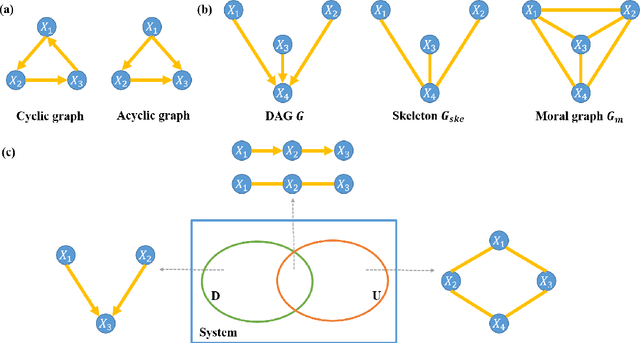
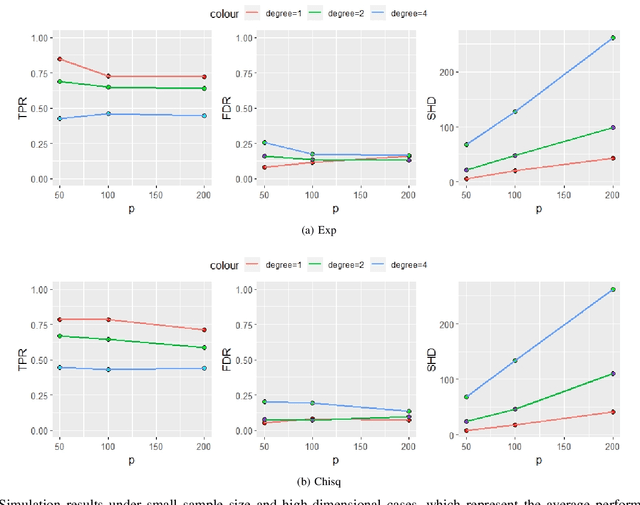
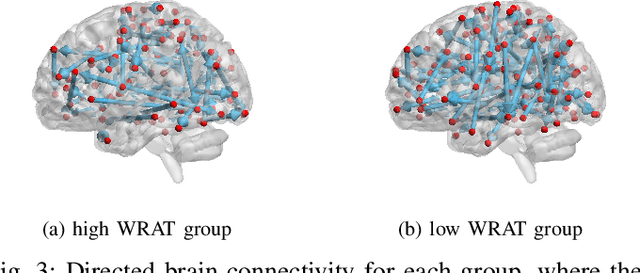
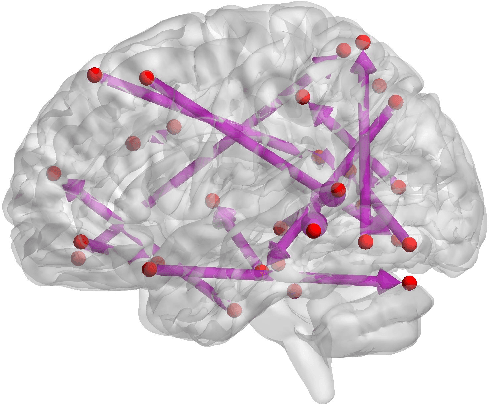
Functional connectivity (FC) has become a primary means of understanding brain functions by identifying brain network interactions and, ultimately, how those interactions produce cognitions. A popular definition of FC is by statistical associations between measured brain regions. However, this could be problematic since the associations can only provide spatial connections but not causal interactions among regions of interests. Hence, it is necessary to study their causal relationship. Directed acyclic graph (DAG) models have been applied in recent FC studies but often encountered problems such as limited sample sizes and large number of variables (namely high-dimensional problems), which lead to both computational difficulty and convergence issues. As a result, the use of DAG models is problematic, where the identification of DAG models in general is nondeterministic polynomial time hard (NP-hard). To this end, we propose a $\psi$-learning incorporated linear non-Gaussian acyclic model ($\psi$-LiNGAM). We use the association model ($\psi$-learning) to facilitate causal inferences and the model works well especially for high-dimensional cases. Our simulation results demonstrate that the proposed method is more robust and accurate than several existing ones in detecting graph structure and direction. We then applied it to the resting state fMRI (rsfMRI) data obtained from the publicly available Philadelphia Neurodevelopmental Cohort (PNC) to study the cognitive variance, which includes 855 individuals aged 8-22 years. Therein, we have identified three types of hub structure: the in-hub, out-hub and sum-hub, which correspond to the centers of receiving, sending and relaying information, respectively. We also detected 16 most important pairs of causal flows. Several of the results have been verified to be biologically significant.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge