Anees Abrol
Tri-Institutional Center for Translational Research in Neuroimaging and Data Science
Hierarchical Spatio-Temporal State-Space Modeling for fMRI Analysis
Aug 23, 2024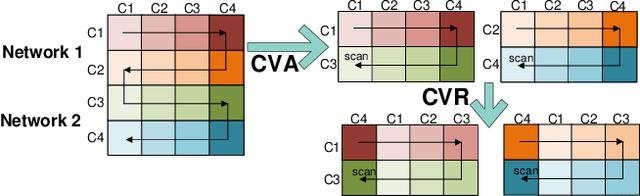
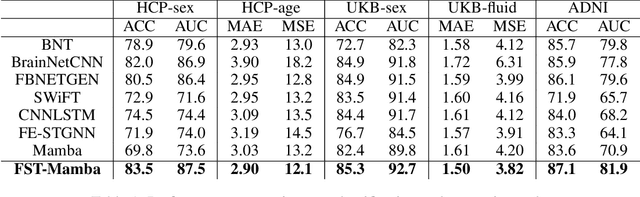
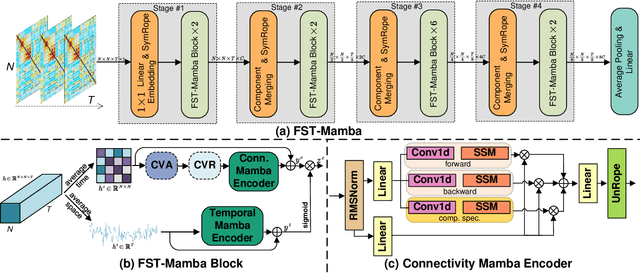

Abstract:Recent advances in deep learning structured state space models, especially the Mamba architecture, have demonstrated remarkable performance improvements while maintaining linear complexity. In this study, we introduce functional spatiotemporal Mamba (FST-Mamba), a Mamba-based model designed for discovering neurological biomarkers using functional magnetic resonance imaging (fMRI). We focus on dynamic functional network connectivity (dFNC) derived from fMRI and propose a hierarchical spatiotemporal Mamba-based network that processes spatial and temporal information separately using Mamba-based encoders. Leveraging the topological uniqueness of the FNC matrix, we introduce a component-wise varied-scale aggregation (CVA) mechanism to aggregate connectivity across individual components within brain networks, enabling the model to capture both inter-component and inter-network information. To better handle the FNC data, we develop a new component-specific scanning order. Additionally, we propose symmetric rotary position encoding (SymRope) to encode the relative positions of each functional connection while considering the symmetric nature of the FNC matrix. Experimental results demonstrate significant improvements in the proposed FST-Mamba model on various brain-based classification and regression tasks. Our work reveals the substantial potential of attention-free sequence modeling in brain discovery.
An interpretable generative multimodal neuroimaging-genomics framework for decoding Alzheimer's disease
Jun 19, 2024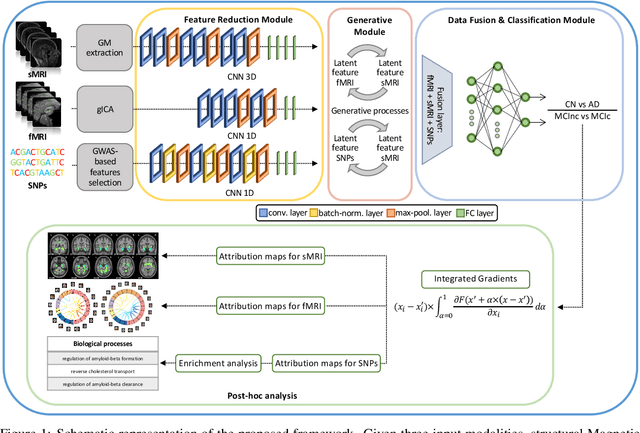

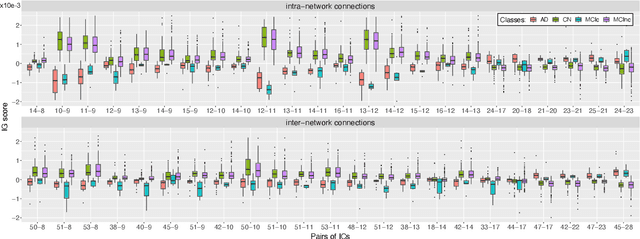

Abstract:Alzheimer's disease (AD) is the most prevalent form of dementia with a progressive decline in cognitive abilities. The AD continuum encompasses a prodormal stage known as Mild Cognitive Impairment (MCI), where patients may either progress to AD or remain stable. In this study, we leveraged structural and functional MRI to investigate the disease-induced grey matter and functional network connectivity changes. Moreover, considering AD's strong genetic component, we introduce SNPs as a third channel. Given such diverse inputs, missing one or more modalities is a typical concern of multimodal methods. We hence propose a novel deep learning-based classification framework where generative module employing Cycle GANs was adopted to impute missing data within the latent space. Additionally, we adopted an Explainable AI method, Integrated Gradients, to extract input features relevance, enhancing our understanding of the learned representations. Two critical tasks were addressed: AD detection and MCI conversion prediction. Experimental results showed that our model was able to reach the SOA in the classification of CN/AD reaching an average test accuracy of $0.926\pm0.02$. For the MCI task, we achieved an average prediction accuracy of $0.711\pm0.01$ using the pre-trained model for CN/AD. The interpretability analysis revealed significant grey matter modulations in cortical and subcortical brain areas well known for their association with AD. Moreover, impairments in sensory-motor and visual resting state network connectivity along the disease continuum, as well as mutations in SNPs defining biological processes linked to amyloid-beta and cholesterol formation clearance and regulation, were identified as contributors to the achieved performance. Overall, our integrative deep learning approach shows promise for AD detection and MCI prediction, while shading light on important biological insights.
Multimodal MRI-based Detection of Amyloid Status in Alzheimer's Disease Continuum
Jun 19, 2024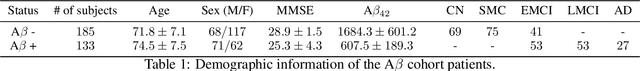
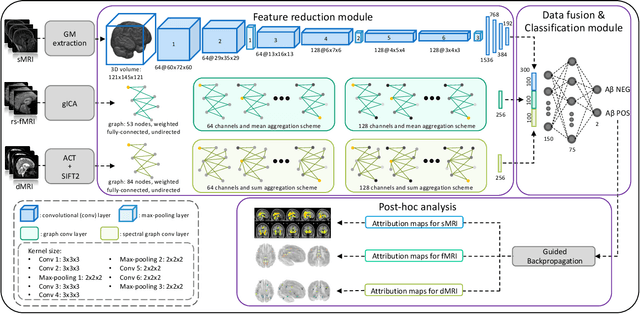

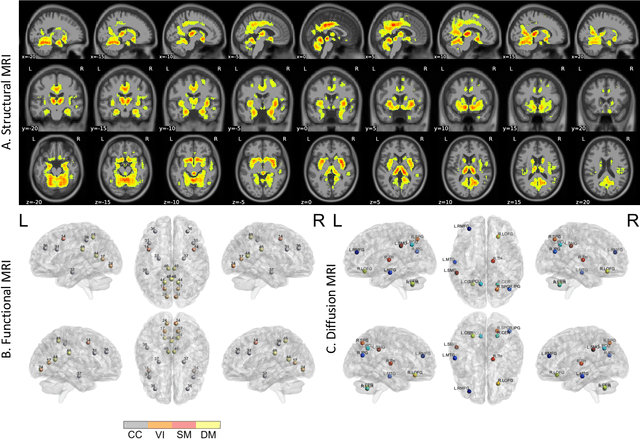
Abstract:Amyloid-$\beta$ (A$\beta$) plaques in conjunction with hyperphosphorylated tau proteins in the form of neurofibrillary tangles are the two neuropathological hallmarks of Alzheimer's disease (AD). In particular, the accumulation of A$\beta$ plaques, as evinced by the A/T/N (amyloid/tau/neurodegeneration) framework, marks the initial stage. Thus, the identification of individuals with A$\beta$ positivity could enable early diagnosis and potentially lead to more effective interventions. Deep learning methods relying mainly on amyloid PET images have been employed to this end. However, PET imaging has some disadvantages, including the need of radiotracers and expensive acquisitions. Hence, in this work, we propose a novel multimodal approach that integrates information from structural, functional, and diffusion MRI data to discriminate A$\beta$ status in the AD continuum. Our method achieved an accuracy of $0.762\pm0.04$. Furthermore, a \textit{post-hoc} explainability analysis (guided backpropagation) was performed to retrieve the brain regions that most influenced the model predictions. This analysis identified some key regions that were common across modalities, some of which were well-established AD-discriminative biomarkers and related to A$\beta$ deposition, such as the hippocampus, thalamus, precuneus, and cingulate gyrus. Hence, our study demonstrates the potential viability of MRI-based characterization of A$\beta$ status, paving the way for further research in this domain.
Cross-Modality Translation with Generative Adversarial Networks to Unveil Alzheimer's Disease Biomarkers
May 08, 2024Abstract:Generative approaches for cross-modality transformation have recently gained significant attention in neuroimaging. While most previous work has focused on case-control data, the application of generative models to disorder-specific datasets and their ability to preserve diagnostic patterns remain relatively unexplored. Hence, in this study, we investigated the use of a generative adversarial network (GAN) in the context of Alzheimer's disease (AD) to generate functional network connectivity (FNC) and T1-weighted structural magnetic resonance imaging data from each other. We employed a cycle-GAN to synthesize data in an unpaired data transition and enhanced the transition by integrating weak supervision in cases where paired data were available. Our findings revealed that our model could offer remarkable capability, achieving a structural similarity index measure (SSIM) of $0.89 \pm 0.003$ for T1s and a correlation of $0.71 \pm 0.004$ for FNCs. Moreover, our qualitative analysis revealed similar patterns between generated and actual data when comparing AD to cognitively normal (CN) individuals. In particular, we observed significantly increased functional connectivity in cerebellar-sensory motor and cerebellar-visual networks and reduced connectivity in cerebellar-subcortical, auditory-sensory motor, sensory motor-visual, and cerebellar-cognitive control networks. Additionally, the T1 images generated by our model showed a similar pattern of atrophy in the hippocampal and other temporal regions of Alzheimer's patients.
Cross-Modal Synthesis of Structural MRI and Functional Connectivity Networks via Conditional ViT-GANs
Sep 15, 2023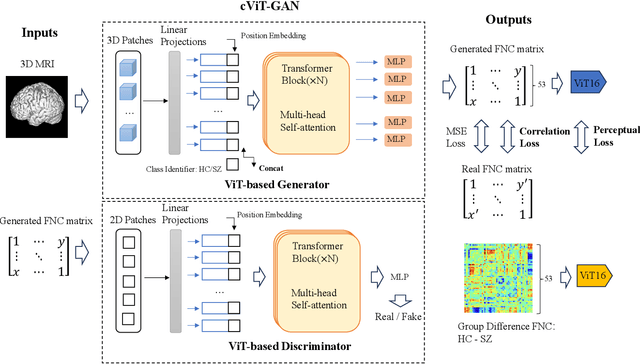
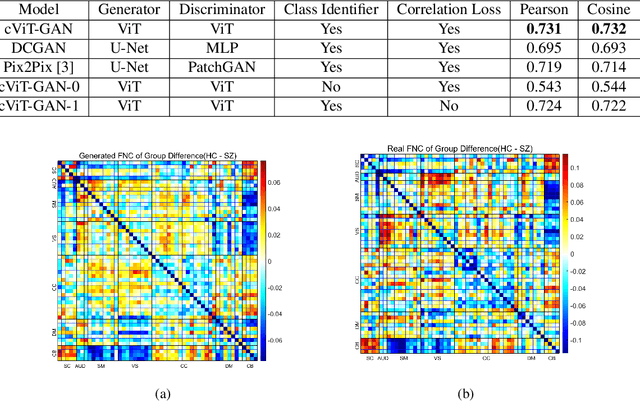
Abstract:The cross-modal synthesis between structural magnetic resonance imaging (sMRI) and functional network connectivity (FNC) is a relatively unexplored area in medical imaging, especially with respect to schizophrenia. This study employs conditional Vision Transformer Generative Adversarial Networks (cViT-GANs) to generate FNC data based on sMRI inputs. After training on a comprehensive dataset that included both individuals with schizophrenia and healthy control subjects, our cViT-GAN model effectively synthesized the FNC matrix for each subject, and then formed a group difference FNC matrix, obtaining a Pearson correlation of 0.73 with the actual FNC matrix. In addition, our FNC visualization results demonstrate significant correlations in particular subcortical brain regions, highlighting the model's capability of capturing detailed structural-functional associations. This performance distinguishes our model from conditional CNN-based GAN alternatives such as Pix2Pix. Our research is one of the first attempts to link sMRI and FNC synthesis, setting it apart from other cross-modal studies that concentrate on T1- and T2-weighted MR images or the fusion of MRI and CT scans.
MultiCrossViT: Multimodal Vision Transformer for Schizophrenia Prediction using Structural MRI and Functional Network Connectivity Data
Nov 12, 2022Abstract:Vision Transformer (ViT) is a pioneering deep learning framework that can address real-world computer vision issues, such as image classification and object recognition. Importantly, ViTs are proven to outperform traditional deep learning models, such as convolutional neural networks (CNNs). Relatively recently, a number of ViT mutations have been transplanted into the field of medical imaging, thereby resolving a variety of critical classification and segmentation challenges, especially in terms of brain imaging data. In this work, we provide a novel multimodal deep learning pipeline, MultiCrossViT, which is capable of analyzing both structural MRI (sMRI) and static functional network connectivity (sFNC) data for the prediction of schizophrenia disease. On a dataset with minimal training subjects, our novel model can achieve an AUC of 0.832. Finally, we visualize multiple brain regions and covariance patterns most relevant to schizophrenia based on the resulting ViT attention maps by extracting features from transformer encoders.
Prediction of Gender from Longitudinal MRI data via Deep Learning on Adolescent Data Reveals Unique Patterns Associated with Brain Structure and Change over a Two-year Period
Sep 15, 2022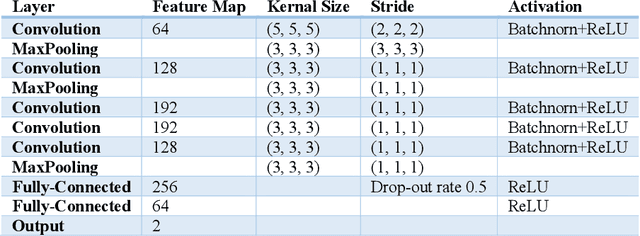
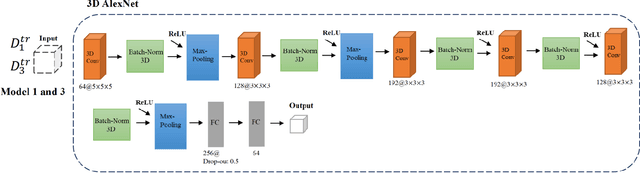
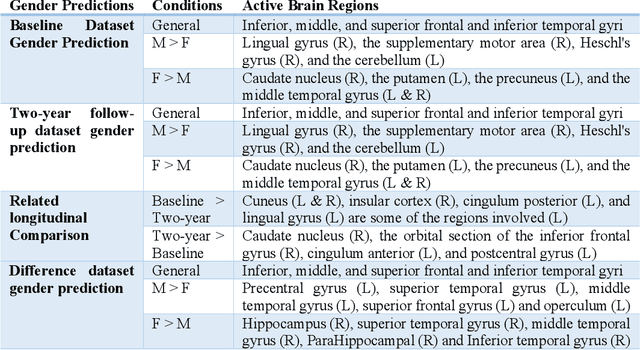
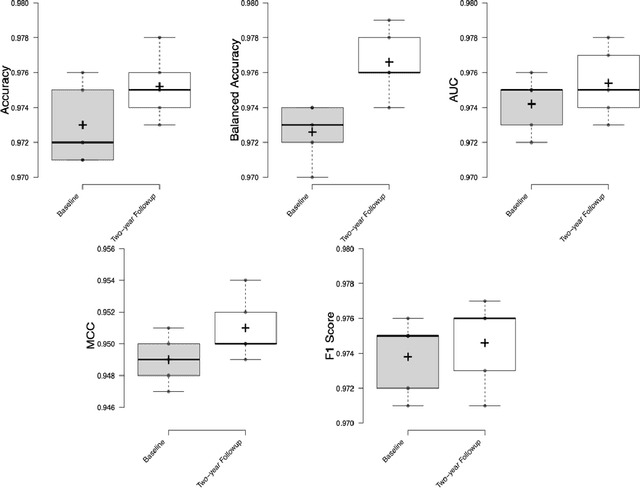
Abstract:Deep learning algorithms for predicting neuroimaging data have shown considerable promise in various applications. Prior work has demonstrated that deep learning models that take advantage of the data's 3D structure can outperform standard machine learning on several learning tasks. However, most prior research in this area has focused on neuroimaging data from adults. Within the Adolescent Brain and Cognitive Development (ABCD) dataset, a large longitudinal development study, we examine structural MRI data to predict gender and identify gender-related changes in brain structure. Results demonstrate that gender prediction accuracy is exceptionally high (>97%) with training epochs >200 and that this accuracy increases with age. Brain regions identified as the most discriminative in the task under study include predominantly frontal areas and the temporal lobe. When evaluating gender predictive changes specific to a two-year increase in age, a broader set of visual, cingulate, and insular regions are revealed. Our findings show a robust gender-related structural brain change pattern, even over a small age range. This suggests that it might be possible to study how the brain changes during adolescence by looking at how these changes are related to different behavioral and environmental factors.
Pipeline-Invariant Representation Learning for Neuroimaging
Aug 27, 2022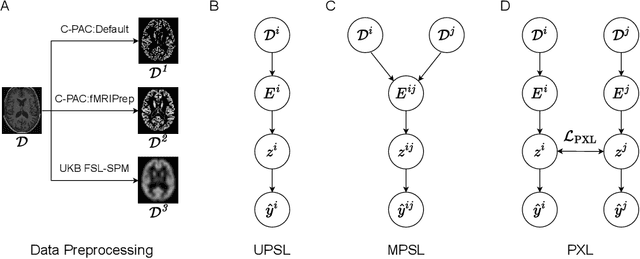

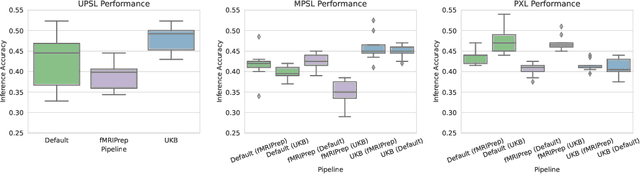
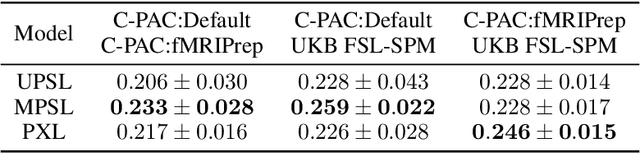
Abstract:Deep learning has been widely applied in neuroimaging, including to predicting brain-phenotype relationships from magnetic resonance imaging (MRI) volumes. MRI data usually requires extensive preprocessing before it is ready for modeling, even via deep learning, in part due to its high dimensionality and heterogeneity. A growing array of MRI preprocessing pipelines have been developed each with its own strengths and limitations. Recent studies have shown that pipeline-related variation may lead to different scientific findings, even when using the identical data. Meanwhile, the machine learning community has emphasized the importance of shifting from model-centric to data-centric approaches given that data quality plays an essential role in deep learning applications. Motivated by this idea, we first evaluate how preprocessing pipeline selection can impact the downstream performance of a supervised learning model. We next propose two pipeline-invariant representation learning methodologies, MPSL and PXL, to improve consistency in classification performance and to capture similar neural network representations between pipeline pairs. Using 2000 human subjects from the UK Biobank dataset, we demonstrate that both models present unique advantages, in particular that MPSL can be used to improve out-of-sample generalization to new pipelines, while PXL can be used to improve predictive performance consistency and representational similarity within a closed pipeline set. These results suggest that our proposed models can be applied to overcome pipeline-related biases and to improve reproducibility in neuroimaging prediction tasks.
Prediction of Progression to Alzheimer's disease with Deep InfoMax
May 01, 2019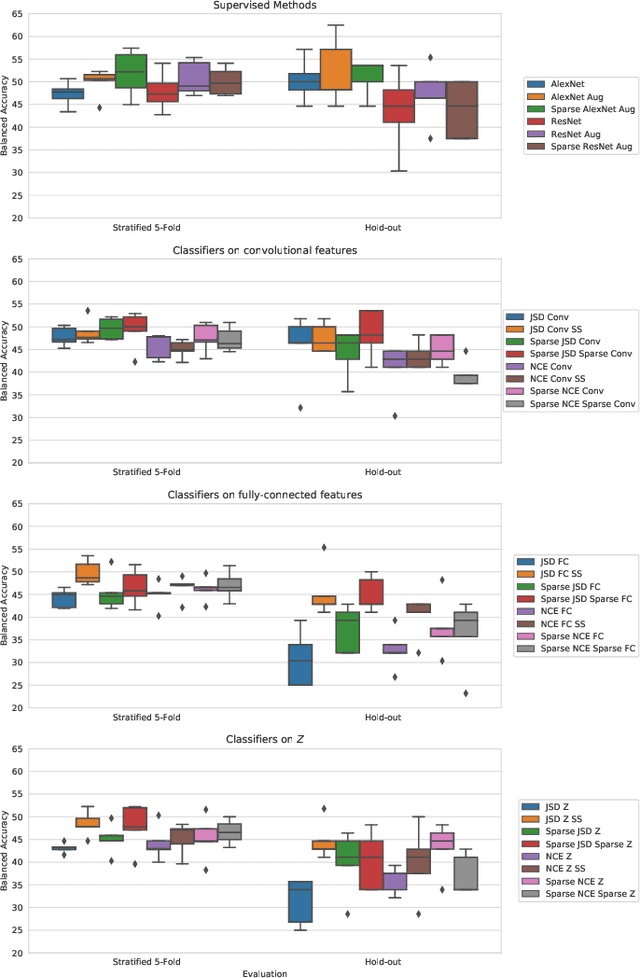
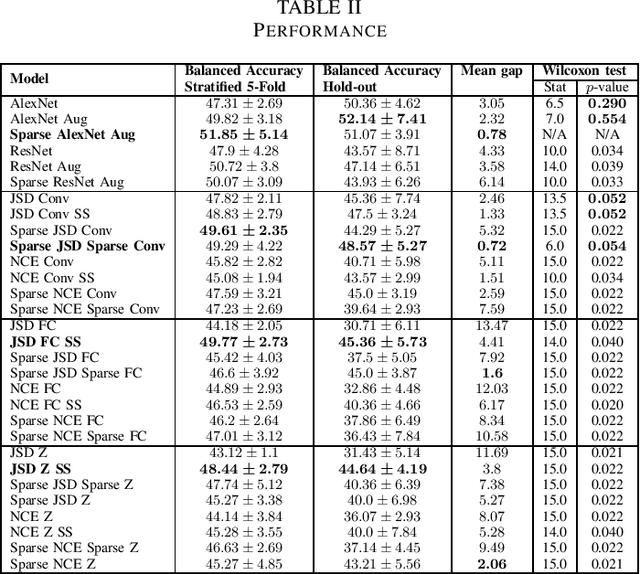
Abstract:Arguably, unsupervised learning plays a crucial role in the majority of algorithms for processing brain imaging. A recently introduced unsupervised approach Deep InfoMax (DIM) is a promising tool for exploring brain structure in a flexible non-linear way. In this paper, we investigate the use of variants of DIM in a setting of progression to Alzheimer's disease in comparison with supervised AlexNet and ResNet inspired convolutional neural networks. As a benchmark, we use a classification task between four groups: patients with stable, and progressive mild cognitive impairment (MCI), with Alzheimer's disease, and healthy controls. Our dataset is comprised of 828 subjects from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database. Our experiments highlight encouraging evidence of the high potential utility of DIM in future neuroimaging studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge