Gregory Kiar
Center for the Developing Brain, Child Mind Institute, New York, NY, USA
A Parameter-efficient Multi-subject Model for Predicting fMRI Activity
Aug 04, 2023Abstract:This is the Algonauts 2023 submission report for team "BlobGPT". Our model consists of a multi-subject linear encoding head attached to a pretrained trunk model. The multi-subject head consists of three components: (1) a shared multi-layer feature projection, (2) shared plus subject-specific low-dimension linear transformations, and (3) a shared PCA fMRI embedding. In this report, we explain these components in more detail and present some experimental results. Our code is available at https://github.com/cmi-dair/algonauts23.
Numerical Stability of DeepGOPlus Inference
Dec 13, 2022Abstract:Convolutional neural networks (CNNs) are currently among the most widely-used neural networks available and achieve state-of-the-art performance for many problems. While originally applied to computer vision tasks, CNNs work well with any data with a spatial relationship, besides images, and have been applied to different fields. However, recent works have highlighted how CNNs, like other deep learning models, are sensitive to noise injection which can jeopardise their performance. This paper quantifies the numerical uncertainty of the floating point arithmetic inaccuracies of the inference stage of DeepGOPlus, a CNN that predicts protein function, in order to determine its numerical stability. In addition, this paper investigates the possibility to use reduced-precision floating point formats for DeepGOPlus inference to reduce memory consumption and latency. This is achieved with Monte Carlo Arithmetic, a technique that experimentally quantifies floating point operation errors and VPREC, a tool that emulates results with customizable floating point precision formats. Focus is placed on the inference stage as it is the main deliverable of the DeepGOPlus model that will be used across environments and therefore most likely be subjected to the most amount of noise. Furthermore, studies have shown that the inference stage is the part of the model which is most disposed to being scaled down in terms of reduced precision. All in all, it has been found that the numerical uncertainty of the DeepGOPlus CNN is very low at its current numerical precision format, but the model cannot currently be reduced to a lower precision that might render it more lightweight.
Pipeline-Invariant Representation Learning for Neuroimaging
Aug 27, 2022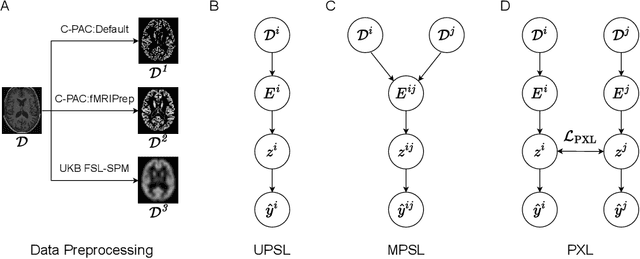

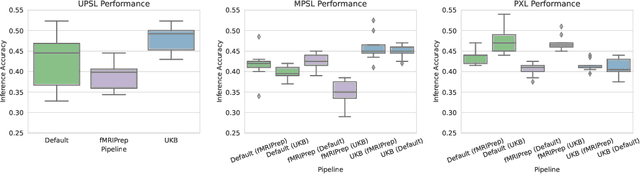
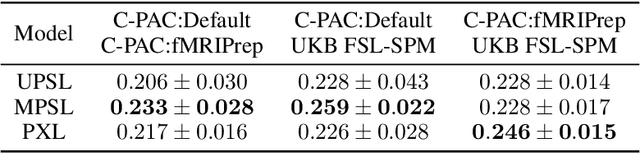
Abstract:Deep learning has been widely applied in neuroimaging, including to predicting brain-phenotype relationships from magnetic resonance imaging (MRI) volumes. MRI data usually requires extensive preprocessing before it is ready for modeling, even via deep learning, in part due to its high dimensionality and heterogeneity. A growing array of MRI preprocessing pipelines have been developed each with its own strengths and limitations. Recent studies have shown that pipeline-related variation may lead to different scientific findings, even when using the identical data. Meanwhile, the machine learning community has emphasized the importance of shifting from model-centric to data-centric approaches given that data quality plays an essential role in deep learning applications. Motivated by this idea, we first evaluate how preprocessing pipeline selection can impact the downstream performance of a supervised learning model. We next propose two pipeline-invariant representation learning methodologies, MPSL and PXL, to improve consistency in classification performance and to capture similar neural network representations between pipeline pairs. Using 2000 human subjects from the UK Biobank dataset, we demonstrate that both models present unique advantages, in particular that MPSL can be used to improve out-of-sample generalization to new pipelines, while PXL can be used to improve predictive performance consistency and representational similarity within a closed pipeline set. These results suggest that our proposed models can be applied to overcome pipeline-related biases and to improve reproducibility in neuroimaging prediction tasks.
Data Augmentation Through Monte Carlo Arithmetic Leads to More Generalizable Classification in Connectomics
Sep 20, 2021Abstract:Machine learning models are commonly applied to human brain imaging datasets in an effort to associate function or structure with behaviour, health, or other individual phenotypes. Such models often rely on low-dimensional maps generated by complex processing pipelines. However, the numerical instabilities inherent to pipelines limit the fidelity of these maps and introduce computational bias. Monte Carlo Arithmetic, a technique for introducing controlled amounts of numerical noise, was used to perturb a structural connectome estimation pipeline, ultimately producing a range of plausible networks for each sample. The variability in the perturbed networks was captured in an augmented dataset, which was then used for an age classification task. We found that resampling brain networks across a series of such numerically perturbed outcomes led to improved performance in all tested classifiers, preprocessing strategies, and dimensionality reduction techniques. Importantly, we find that this benefit does not hinge on a large number of perturbations, suggesting that even minimally perturbing a dataset adds meaningful variance which can be captured in the subsequently designed models.
A Recommender System for Scientific Datasets and Analysis Pipelines
Aug 20, 2021



Abstract:Scientific datasets and analysis pipelines are increasingly being shared publicly in the interest of open science. However, mechanisms are lacking to reliably identify which pipelines and datasets can appropriately be used together. Given the increasing number of high-quality public datasets and pipelines, this lack of clear compatibility threatens the findability and reusability of these resources. We investigate the feasibility of a collaborative filtering system to recommend pipelines and datasets based on provenance records from previous executions. We evaluate our system using datasets and pipelines extracted from the Canadian Open Neuroscience Platform, a national initiative for open neuroscience. The recommendations provided by our system (AUC$=0.83$) are significantly better than chance and outperform recommendations made by domain experts using their previous knowledge as well as pipeline and dataset descriptions (AUC$=0.63$). In particular, domain experts often neglect low-level technical aspects of a pipeline-dataset interaction, such as the level of pre-processing, which are captured by a provenance-based system. We conclude that provenance-based pipeline and dataset recommenders are feasible and beneficial to the sharing and usage of open-science resources. Future work will focus on the collection of more comprehensive provenance traces, and on deploying the system in production.
Accurate simulation of operating system updates in neuroimaging using Monte-Carlo arithmetic
Aug 06, 2021



Abstract:Operating system (OS) updates introduce numerical perturbations that impact the reproducibility of computational pipelines. In neuroimaging, this has important practical implications on the validity of computational results, particularly when obtained in systems such as high-performance computing clusters where the experimenter does not control software updates. We present a framework to reproduce the variability induced by OS updates in controlled conditions. We hypothesize that OS updates impact computational pipelines mainly through numerical perturbations originating in mathematical libraries, which we simulate using Monte-Carlo arithmetic in a framework called "fuzzy libmath" (FL). We applied this methodology to pre-processing pipelines of the Human Connectome Project, a flagship open-data project in neuroimaging. We found that FL-perturbed pipelines accurately reproduce the variability induced by OS updates and that this similarity is only mildly dependent on simulation parameters. Importantly, we also found between-subject differences were preserved in both cases, though the between-run variability was of comparable magnitude for both FL and OS perturbations. We found the numerical precision in the HCP pre-processed images to be relatively low, with less than 8 significant bits among the 24 available, which motivates further investigation of the numerical stability of components in the tested pipeline. Overall, our results establish that FL accurately simulates results variability due to OS updates, and is a practical framework to quantify numerical uncertainty in neuroimaging.
Reducing numerical precision preserves classification accuracy in Mondrian Forests
Jun 28, 2021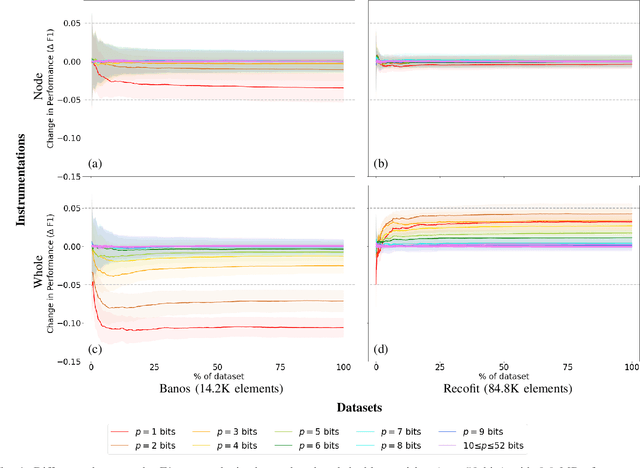
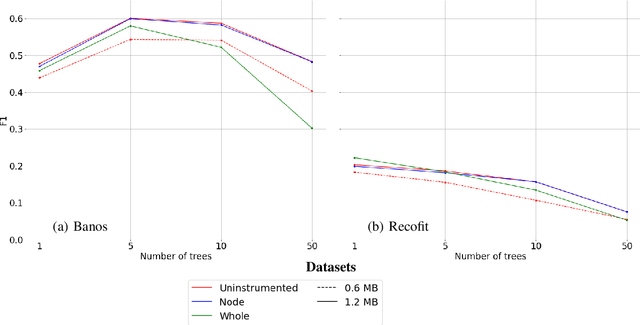

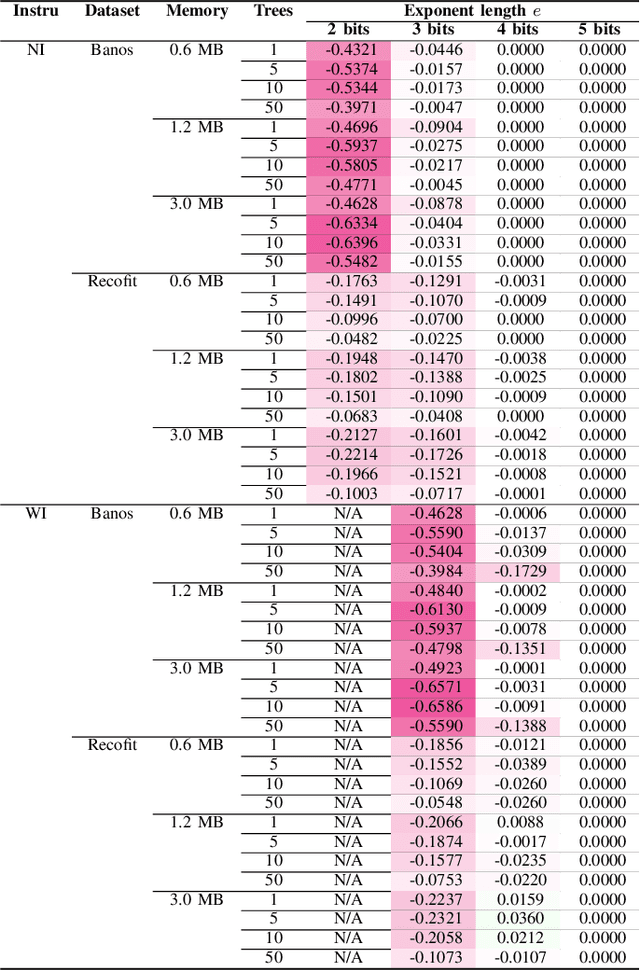
Abstract:Mondrian Forests are a powerful data stream classification method, but their large memory footprint makes them ill-suited for low-resource platforms such as connected objects. We explored using reduced-precision floating-point representations to lower memory consumption and evaluated its effect on classification performance. We applied the Mondrian Forest implementation provided by OrpailleCC, a C++ collection of data stream algorithms, to two canonical datasets in human activity recognition: Recofit and Banos \emph{et al}. Results show that the precision of floating-point values used by tree nodes can be reduced from 64 bits to 8 bits with no significant difference in F1 score. In some cases, reduced precision was shown to improve classification performance, presumably due to its regularization effect. We conclude that numerical precision is a relevant hyperparameter in the Mondrian Forest, and that commonly-used double precision values may not be necessary for optimal performance. Future work will evaluate the generalizability of these findings to other data stream classifiers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge