Tristan Glatard
Department of Computer Science and Software Engineering, Concordia University, Montréal, QC, Canada
Conservative & Aggressive NaNs Accelerate U-Nets for Neuroimaging
Jan 23, 2026Abstract:Deep learning models for neuroimaging increasingly rely on large architectures, making efficiency a persistent concern despite advances in hardware. Through an analysis of numerical uncertainty of convolutional neural networks (CNNs), we observe that many operations are applied to values dominated by numerical noise and have negligible influence on model outputs. In some models, up to two-thirds of convolution operations appear redundant. We introduce Conservative & Aggressive NaNs, two novel variants of max pooling and unpooling that identify numerically unstable voxels and replace them with NaNs, allowing subsequent layers to skip computations on irrelevant data. Both methods are implemented within PyTorch and require no architectural changes. We evaluate these approaches on four CNN models spanning neuroimaging and image classification tasks. For inputs containing at least 50% NaNs, we observe consistent runtime improvements; for data with more than two-thirds NaNs )common in several neuroimaging settings) we achieve an average inference speedup of 1.67x. Conservative NaNs reduces convolution operations by an average of 30% across models and datasets, with no measurable performance degradation, and can skip up to 64.64% of convolutions in specific layers. Aggressive NaNs can skip up to 69.30% of convolutions but may occasionally affect performance. Overall, these methods demonstrate that numerical uncertainty can be exploited to reduce redundant computation and improve inference efficiency in CNNs.
Uncertain but Useful: Leveraging CNN Variability into Data Augmentation
Sep 05, 2025Abstract:Deep learning (DL) is rapidly advancing neuroimaging by achieving state-of-the-art performance with reduced computation times. Yet the numerical stability of DL models -- particularly during training -- remains underexplored. While inference with DL is relatively stable, training introduces additional variability primarily through iterative stochastic optimization. We investigate this training-time variability using FastSurfer, a CNN-based whole-brain segmentation pipeline. Controlled perturbations are introduced via floating point perturbations and random seeds. We find that: (i) FastSurfer exhibits higher variability compared to that of a traditional neuroimaging pipeline, suggesting that DL inherits and is particularly susceptible to sources of instability present in its predecessors; (ii) ensembles generated with perturbations achieve performance similar to an unperturbed baseline; and (iii) variability effectively produces ensembles of numerical model families that can be repurposed for downstream applications. As a proof of concept, we demonstrate that numerical ensembles can be used as a data augmentation strategy for brain age regression. These findings position training-time variability not only as a reproducibility concern but also as a resource that can be harnessed to improve robustness and enable new applications in neuroimaging.
Numerical Uncertainty in Linear Registration: An Experimental Study
Aug 01, 2025Abstract:While linear registration is a critical step in MRI preprocessing pipelines, its numerical uncertainty is understudied. Using Monte-Carlo Arithmetic (MCA) simulations, we assessed the most commonly used linear registration tools within major software packages (SPM, FSL, and ANTs) across multiple image similarity measures, two brain templates, and both healthy control (HC, n=50) and Parkinson's Disease (PD, n=50) cohorts. Our findings highlight the influence of linear registration tools and similarity measures on numerical stability. Among the evaluated tools and with default similarity measures, SPM exhibited the highest stability. FSL and ANTs showed greater and similar ranges of variability, with ANTs demonstrating particular sensitivity to numerical perturbations that occasionally led to registration failure. Furthermore, no significant differences were observed between healthy and PD cohorts, suggesting that numerical stability analyses obtained with healthy subjects may generalise to clinical populations. Finally, we also demonstrated how numerical uncertainty measures may support automated quality control (QC) of linear registration results. Overall, our experimental results characterize the numerical stability of linear registration experimentally and can serve as a basis for future uncertainty analyses.
Scaling up ridge regression for brain encoding in a massive individual fMRI dataset
Mar 28, 2024



Abstract:Brain encoding with neuroimaging data is an established analysis aimed at predicting human brain activity directly from complex stimuli features such as movie frames. Typically, these features are the latent space representation from an artificial neural network, and the stimuli are image, audio, or text inputs. Ridge regression is a popular prediction model for brain encoding due to its good out-of-sample generalization performance. However, training a ridge regression model can be highly time-consuming when dealing with large-scale deep functional magnetic resonance imaging (fMRI) datasets that include many space-time samples of brain activity. This paper evaluates different parallelization techniques to reduce the training time of brain encoding with ridge regression on the CNeuroMod Friends dataset, one of the largest deep fMRI resource currently available. With multi-threading, our results show that the Intel Math Kernel Library (MKL) significantly outperforms the OpenBLAS library, being 1.9 times faster using 32 threads on a single machine. We then evaluated the Dask multi-CPU implementation of ridge regression readily available in scikit-learn (MultiOutput), and we proposed a new "batch" version of Dask parallelization, motivated by a time complexity analysis. In line with our theoretical analysis, MultiOutput parallelization was found to be impractical, i.e., slower than multi-threading on a single machine. In contrast, the Batch-MultiOutput regression scaled well across compute nodes and threads, providing speed-ups of up to 33 times with 8 compute nodes and 32 threads compared to a single-threaded scikit-learn execution. Batch parallelization using Dask thus emerges as a scalable approach for brain encoding with ridge regression on high-performance computing systems using scikit-learn and large fMRI datasets.
Classification of Anomalies in Telecommunication Network KPI Time Series
Aug 30, 2023

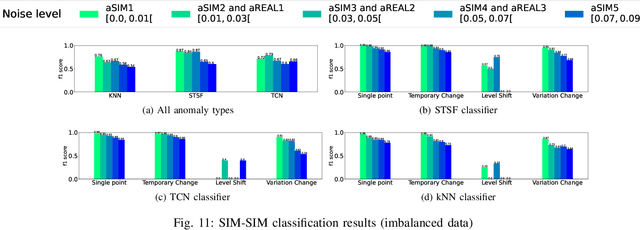

Abstract:The increasing complexity and scale of telecommunication networks have led to a growing interest in automated anomaly detection systems. However, the classification of anomalies detected on network Key Performance Indicators (KPI) has received less attention, resulting in a lack of information about anomaly characteristics and classification processes. To address this gap, this paper proposes a modular anomaly classification framework. The framework assumes separate entities for the anomaly classifier and the detector, allowing for a distinct treatment of anomaly detection and classification tasks on time series. The objectives of this study are (1) to develop a time series simulator that generates synthetic time series resembling real-world network KPI behavior, (2) to build a detection model to identify anomalies in the time series, (3) to build classification models that accurately categorize detected anomalies into predefined classes (4) to evaluate the classification framework performance on simulated and real-world network KPI time series. This study has demonstrated the good performance of the anomaly classification models trained on simulated anomalies when applied to real-world network time series data.
Numerical Uncertainty of Convolutional Neural Networks Inference for Structural Brain MRI Analysis
Aug 03, 2023Abstract:This paper investigates the numerical uncertainty of Convolutional Neural Networks (CNNs) inference for structural brain MRI analysis. It applies Random Rounding -- a stochastic arithmetic technique -- to CNN models employed in non-linear registration (SynthMorph) and whole-brain segmentation (FastSurfer), and compares the resulting numerical uncertainty to the one measured in a reference image-processing pipeline (FreeSurfer recon-all). Results obtained on 32 representative subjects show that CNN predictions are substantially more accurate numerically than traditional image-processing results (non-linear registration: 19 vs 13 significant bits on average; whole-brain segmentation: 0.99 vs 0.92 S{\o}rensen-Dice score on average), which suggests a better reproducibility of CNN results across execution environments.
Numerical Stability of DeepGOPlus Inference
Dec 13, 2022Abstract:Convolutional neural networks (CNNs) are currently among the most widely-used neural networks available and achieve state-of-the-art performance for many problems. While originally applied to computer vision tasks, CNNs work well with any data with a spatial relationship, besides images, and have been applied to different fields. However, recent works have highlighted how CNNs, like other deep learning models, are sensitive to noise injection which can jeopardise their performance. This paper quantifies the numerical uncertainty of the floating point arithmetic inaccuracies of the inference stage of DeepGOPlus, a CNN that predicts protein function, in order to determine its numerical stability. In addition, this paper investigates the possibility to use reduced-precision floating point formats for DeepGOPlus inference to reduce memory consumption and latency. This is achieved with Monte Carlo Arithmetic, a technique that experimentally quantifies floating point operation errors and VPREC, a tool that emulates results with customizable floating point precision formats. Focus is placed on the inference stage as it is the main deliverable of the DeepGOPlus model that will be used across environments and therefore most likely be subjected to the most amount of noise. Furthermore, studies have shown that the inference stage is the part of the model which is most disposed to being scaled down in terms of reduced precision. All in all, it has been found that the numerical uncertainty of the DeepGOPlus CNN is very low at its current numerical precision format, but the model cannot currently be reduced to a lower precision that might render it more lightweight.
Dynamic Ensemble Size Adjustment for Memory Constrained Mondrian Forest
Oct 11, 2022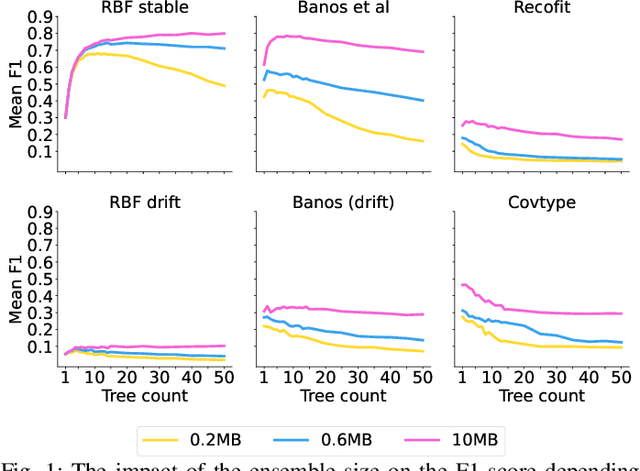
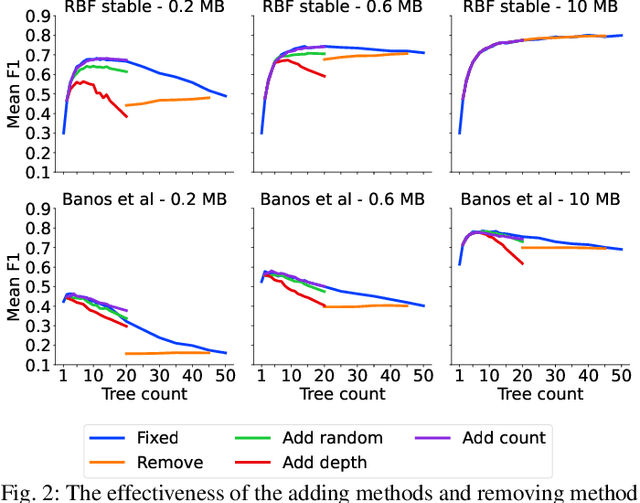
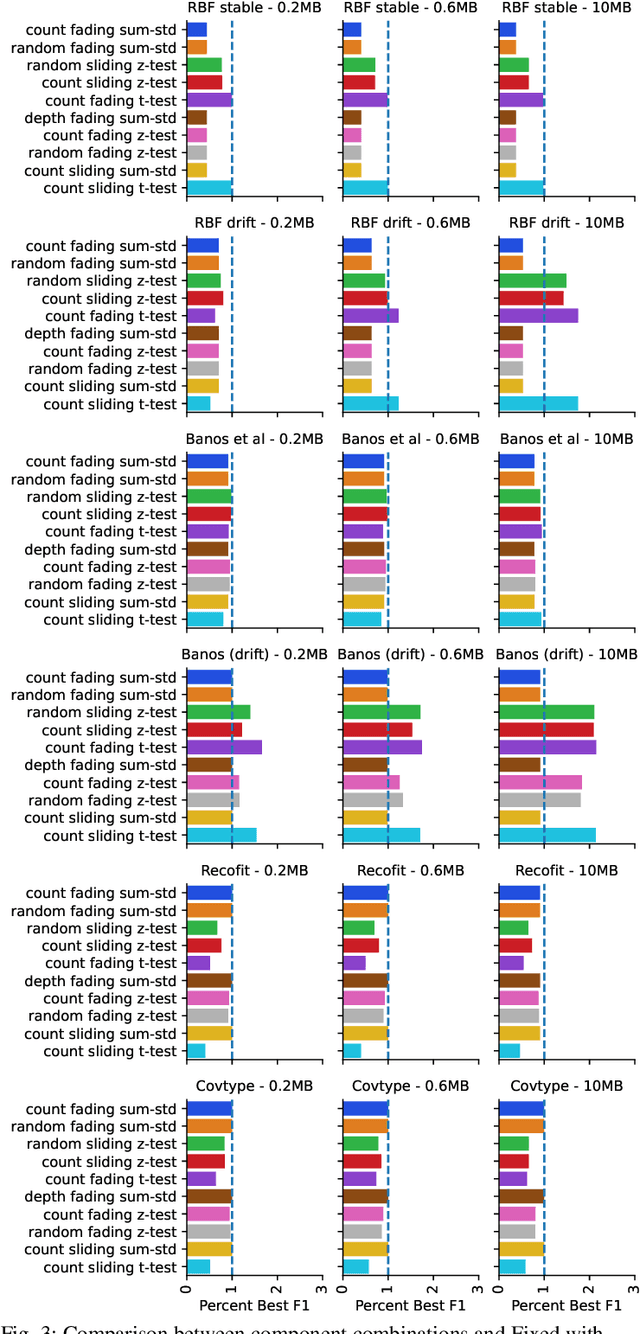
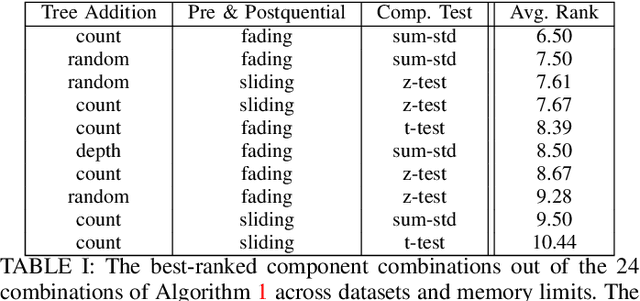
Abstract:Supervised learning algorithms generally assume the availability of enough memory to store data models during the training and test phases. However, this assumption is unrealistic when data comes in the form of infinite data streams, or when learning algorithms are deployed on devices with reduced amounts of memory. Such memory constraints impact the model behavior and assumptions. In this paper, we show that under memory constraints, increasing the size of a tree-based ensemble classifier can worsen its performance. In particular, we experimentally show the existence of an optimal ensemble size for a memory-bounded Mondrian forest on data streams and we design an algorithm to guide the forest toward that optimal number by using an estimation of overfitting. We tested different variations for this algorithm on a variety of real and simulated datasets, and we conclude that our method can achieve up to 95% of the performance of an optimally-sized Mondrian forest for stable datasets, and can even outperform it for datasets with concept drifts. All our methods are implemented in the OrpailleCC open-source library and are ready to be used on embedded systems and connected objects.
Mondrian Forest for Data Stream Classification Under Memory Constraints
May 12, 2022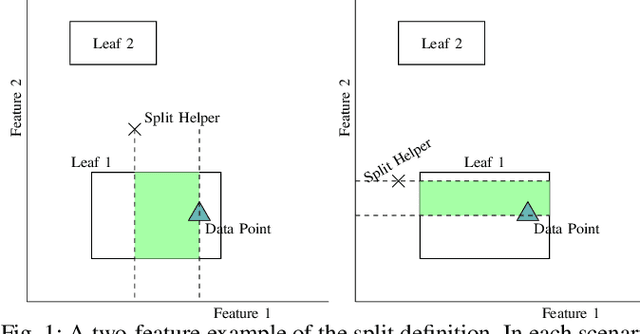
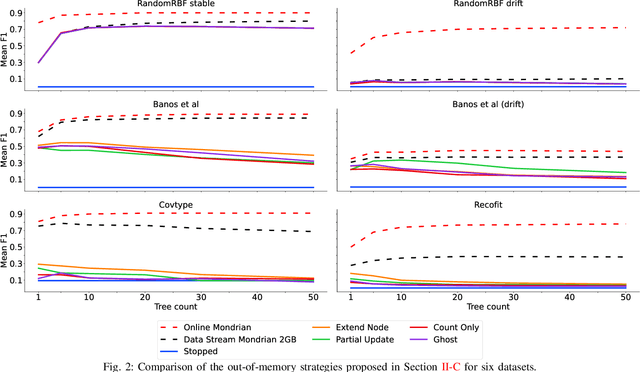
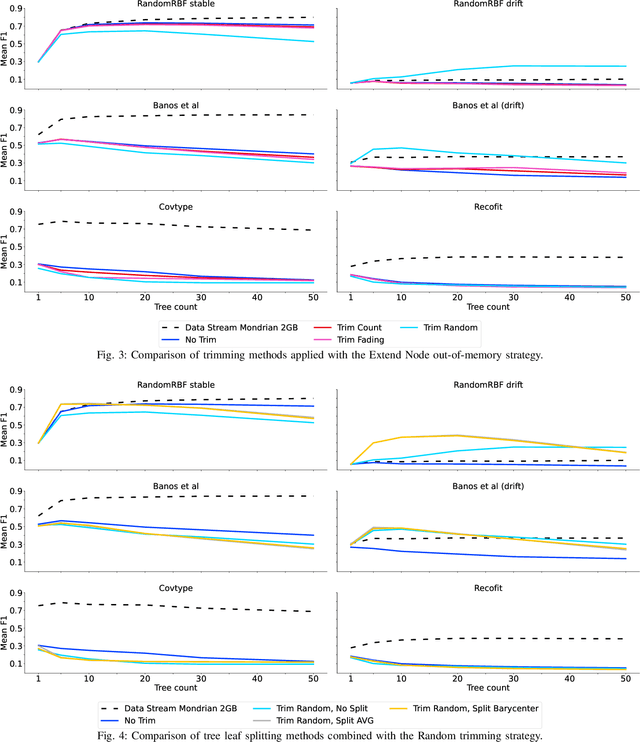
Abstract:Supervised learning algorithms generally assume the availability of enough memory to store their data model during the training and test phases. However, in the Internet of Things, this assumption is unrealistic when data comes in the form of infinite data streams, or when learning algorithms are deployed on devices with reduced amounts of memory. In this paper, we adapt the online Mondrian forest classification algorithm to work with memory constraints on data streams. In particular, we design five out-of-memory strategies to update Mondrian trees with new data points when the memory limit is reached. Moreover, we design trimming mechanisms to make Mondrian trees more robust to concept drifts under memory constraints. We evaluate our algorithms on a variety of real and simulated datasets, and we conclude with recommendations on their use in different situations: the Extend Node strategy appears as the best out-of-memory strategy in all configurations, whereas different trimming mechanisms should be adopted depending on whether a concept drift is expected. All our methods are implemented in the OrpailleCC open-source library and are ready to be used on embedded systems and connected objects.
Data Augmentation Through Monte Carlo Arithmetic Leads to More Generalizable Classification in Connectomics
Sep 20, 2021Abstract:Machine learning models are commonly applied to human brain imaging datasets in an effort to associate function or structure with behaviour, health, or other individual phenotypes. Such models often rely on low-dimensional maps generated by complex processing pipelines. However, the numerical instabilities inherent to pipelines limit the fidelity of these maps and introduce computational bias. Monte Carlo Arithmetic, a technique for introducing controlled amounts of numerical noise, was used to perturb a structural connectome estimation pipeline, ultimately producing a range of plausible networks for each sample. The variability in the perturbed networks was captured in an augmented dataset, which was then used for an age classification task. We found that resampling brain networks across a series of such numerically perturbed outcomes led to improved performance in all tested classifiers, preprocessing strategies, and dimensionality reduction techniques. Importantly, we find that this benefit does not hinge on a large number of perturbations, suggesting that even minimally perturbing a dataset adds meaningful variance which can be captured in the subsequently designed models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge