Pipeline-Invariant Representation Learning for Neuroimaging
Paper and Code
Aug 27, 2022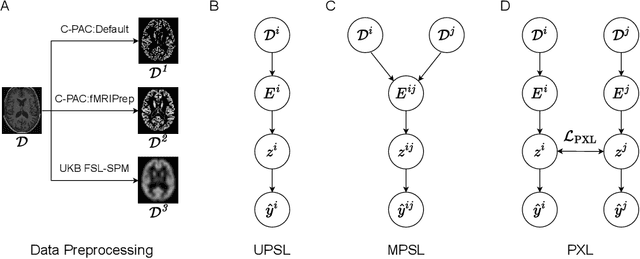

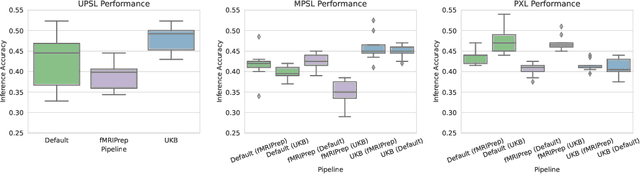
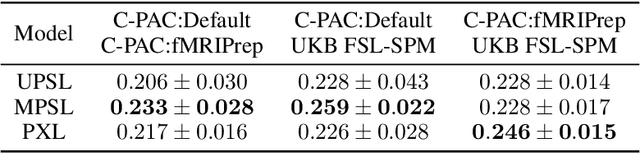
Deep learning has been widely applied in neuroimaging, including to predicting brain-phenotype relationships from magnetic resonance imaging (MRI) volumes. MRI data usually requires extensive preprocessing before it is ready for modeling, even via deep learning, in part due to its high dimensionality and heterogeneity. A growing array of MRI preprocessing pipelines have been developed each with its own strengths and limitations. Recent studies have shown that pipeline-related variation may lead to different scientific findings, even when using the identical data. Meanwhile, the machine learning community has emphasized the importance of shifting from model-centric to data-centric approaches given that data quality plays an essential role in deep learning applications. Motivated by this idea, we first evaluate how preprocessing pipeline selection can impact the downstream performance of a supervised learning model. We next propose two pipeline-invariant representation learning methodologies, MPSL and PXL, to improve consistency in classification performance and to capture similar neural network representations between pipeline pairs. Using 2000 human subjects from the UK Biobank dataset, we demonstrate that both models present unique advantages, in particular that MPSL can be used to improve out-of-sample generalization to new pipelines, while PXL can be used to improve predictive performance consistency and representational similarity within a closed pipeline set. These results suggest that our proposed models can be applied to overcome pipeline-related biases and to improve reproducibility in neuroimaging prediction tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge