Sergey Plis
Tri-Institutional Center for Translational Research in Neuroimaging and Data Science
Learning ECG Representations via Poly-Window Contrastive Learning
Aug 21, 2025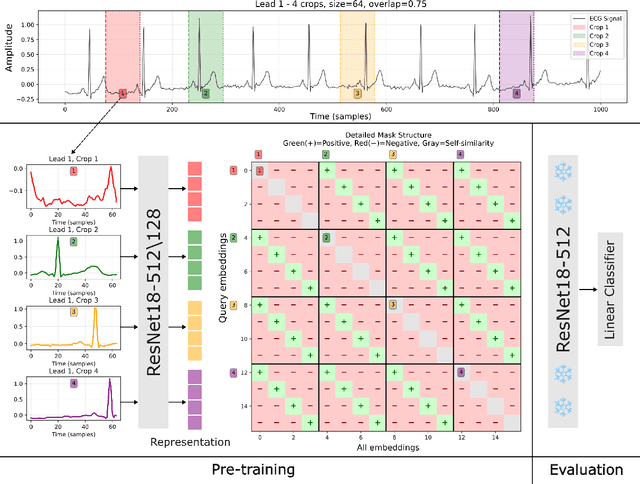
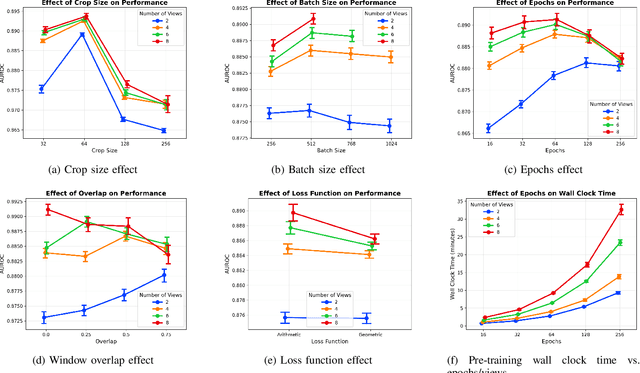
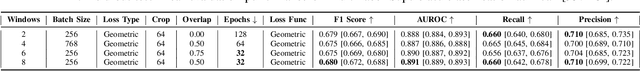

Abstract:Electrocardiogram (ECG) analysis is foundational for cardiovascular disease diagnosis, yet the performance of deep learning models is often constrained by limited access to annotated data. Self-supervised contrastive learning has emerged as a powerful approach for learning robust ECG representations from unlabeled signals. However, most existing methods generate only pairwise augmented views and fail to leverage the rich temporal structure of ECG recordings. In this work, we present a poly-window contrastive learning framework. We extract multiple temporal windows from each ECG instance to construct positive pairs and maximize their agreement via statistics. Inspired by the principle of slow feature analysis, our approach explicitly encourages the model to learn temporally invariant and physiologically meaningful features that persist across time. We validate our approach through extensive experiments and ablation studies on the PTB-XL dataset. Our results demonstrate that poly-window contrastive learning consistently outperforms conventional two-view methods in multi-label superclass classification, achieving higher AUROC (0.891 vs. 0.888) and F1 scores (0.680 vs. 0.679) while requiring up to four times fewer pre-training epochs (32 vs. 128) and 14.8% in total wall clock pre-training time reduction. Despite processing multiple windows per sample, we achieve a significant reduction in the number of training epochs and total computation time, making our method practical for training foundational models. Through extensive ablations, we identify optimal design choices and demonstrate robustness across various hyperparameters. These findings establish poly-window contrastive learning as a highly efficient and scalable paradigm for automated ECG analysis and provide a promising general framework for self-supervised representation learning in biomedical time-series data.
Change of Thought: Adaptive Test-Time Computation
Jul 17, 2025Abstract:Transformers evaluated in a single, fixed-depth pass are provably limited in expressive power to the constant-depth circuit class TC0. Running a Transformer autoregressively removes that ceiling -- first in next-token prediction and, more recently, in chain-of-thought reasoning. Both regimes rely on feedback loops that decode internal states into tokens only to re-encode them in subsequent steps. While this "thinking aloud" mirrors human reasoning, biological brains iterate without externalising intermediate states as language. To boost the expressive power of encoder Transformers without resorting to token-level autoregression, we introduce the SELF-Transformer: an encoder layer that iteratively refines its own attention weights to a fixed point. Instead of producing -- in one pass -- the alignment matrix that remixes the input sequence, the SELF-Transformer iteratively updates that matrix internally, scaling test-time computation with input difficulty. This adaptivity yields up to 20\% accuracy gains on encoder-style benchmarks without increasing parameter count, demonstrating that input-adaptive alignment at test time offers substantial benefits for only a modest extra compute budget. Self-Transformers thus recover much of the expressive power of iterative reasoning while preserving the simplicity of pure encoder architectures.
MindGrab for BrainChop: Fast and Accurate Skull Stripping for Command Line and Browser
Jun 13, 2025Abstract:We developed MindGrab, a parameter- and memory-efficient deep fully-convolutional model for volumetric skull-stripping in head images of any modality. Its architecture, informed by a spectral interpretation of dilated convolutions, was trained exclusively on modality-agnostic synthetic data. MindGrab was evaluated on a retrospective dataset of 606 multimodal adult-brain scans (T1, T2, DWI, MRA, PDw MRI, EPI, CT, PET) sourced from the SynthStrip dataset. Performance was benchmarked against SynthStrip, ROBEX, and BET using Dice scores, with Wilcoxon signed-rank significance tests. MindGrab achieved a mean Dice score of 95.9 with standard deviation (SD) 1.6 across modalities, significantly outperforming classical methods (ROBEX: 89.1 SD 7.7, P < 0.05; BET: 85.2 SD 14.4, P < 0.05). Compared to SynthStrip (96.5 SD 1.1, P=0.0352), MindGrab delivered equivalent or superior performance in nearly half of the tested scenarios, with minor differences (<3% Dice) in the others. MindGrab utilized 95% fewer parameters (146,237 vs. 2,566,561) than SynthStrip. This efficiency yielded at least 2x faster inference, 50% lower memory usage on GPUs, and enabled exceptional performance (e.g., 10-30x speedup, and up to 30x memory reduction) and accessibility on a wider range of hardware, including systems without high-end GPUs. MindGrab delivers state-of-the-art accuracy with dramatically lower resource demands, supported in brainchop-cli (https://pypi.org/project/brainchop/) and at brainchop.org.
Causal Graph Recovery in Neuroimaging through Answer Set Programming
Jun 10, 2025Abstract:Learning graphical causal structures from time series data presents significant challenges, especially when the measurement frequency does not match the causal timescale of the system. This often leads to a set of equally possible underlying causal graphs due to information loss from sub-sampling (i.e., not observing all possible states of the system throughout time). Our research addresses this challenge by incorporating the effects of sub-sampling in the derivation of causal graphs, resulting in more accurate and intuitive outcomes. We use a constraint optimization approach, specifically answer set programming (ASP), to find the optimal set of answers. ASP not only identifies the most probable underlying graph, but also provides an equivalence class of possible graphs for expert selection. In addition, using ASP allows us to leverage graph theory to further prune the set of possible solutions, yielding a smaller, more accurate answer set significantly faster than traditional approaches. We validate our approach on both simulated data and empirical structural brain connectivity, and demonstrate its superiority over established methods in these experiments. We further show how our method can be used as a meta-approach on top of established methods to obtain, on average, 12% improvement in F1 score. In addition, we achieved state of the art results in terms of precision and recall of reconstructing causal graph from sub-sampled time series data. Finally, our method shows robustness to varying degrees of sub-sampling on realistic simulations, whereas other methods perform worse for higher rates of sub-sampling.
State-of-the-Art Stroke Lesion Segmentation at 1/1000th of Parameters
Mar 07, 2025



Abstract:Efficient and accurate whole-brain lesion segmentation remains a challenge in medical image analysis. In this work, we revisit MeshNet, a parameter-efficient segmentation model, and introduce a novel multi-scale dilation pattern with an encoder-decoder structure. This innovation enables capturing broad contextual information and fine-grained details without traditional downsampling, upsampling, or skip-connections. Unlike previous approaches processing subvolumes or slices, we operate directly on whole-brain $256^3$ MRI volumes. Evaluations on the Aphasia Recovery Cohort (ARC) dataset demonstrate that MeshNet achieves superior or comparable DICE scores to state-of-the-art architectures such as MedNeXt and U-MAMBA at 1/1000th of parameters. Our results validate MeshNet's strong balance of efficiency and performance, making it particularly suitable for resource-limited environments such as web-based applications and opening new possibilities for the widespread deployment of advanced medical image analysis tools.
Rethinking Functional Brain Connectome Analysis: Do Graph Deep Learning Models Help?
Jan 28, 2025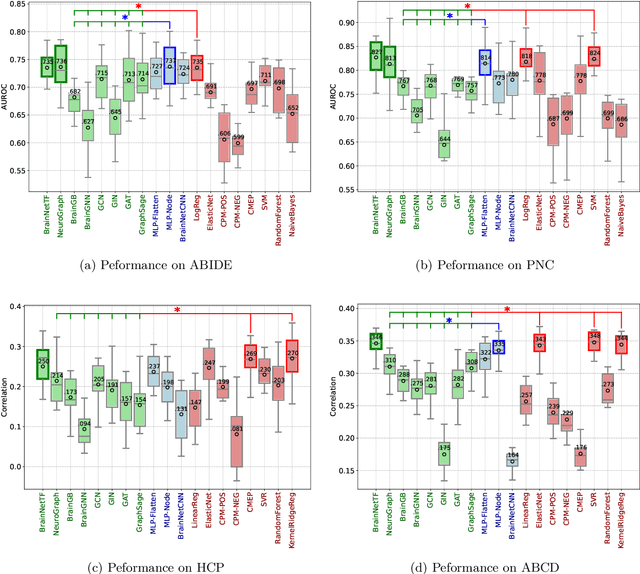
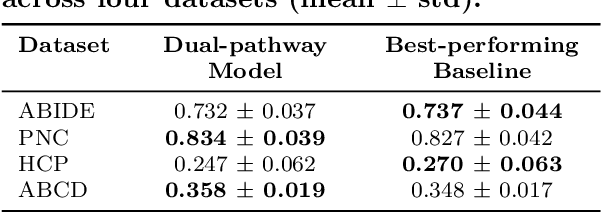
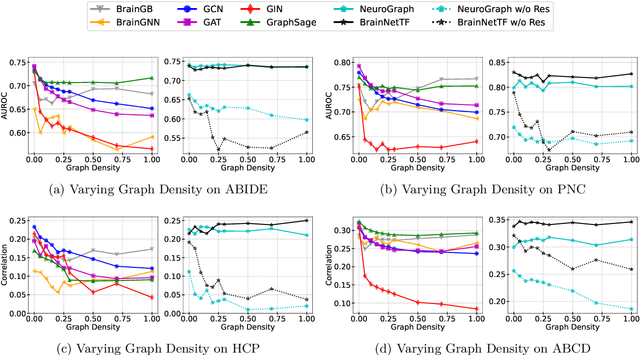
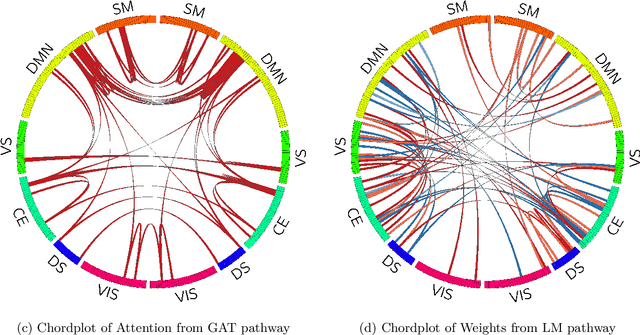
Abstract:Functional brain connectome is crucial for deciphering the neural mechanisms underlying cognitive functions and neurological disorders. Graph deep learning models have recently gained tremendous popularity in this field. However, their actual effectiveness in modeling the brain connectome remains unclear. In this study, we re-examine graph deep learning models based on four large-scale neuroimaging studies encompassing diverse cognitive and clinical outcomes. Surprisingly, we find that the message aggregation mechanism, a hallmark of graph deep learning models, does not help with predictive performance as typically assumed, but rather consistently degrades it. To address this issue, we propose a hybrid model combining a linear model with a graph attention network through dual pathways, achieving robust predictions and enhanced interpretability by revealing both localized and global neural connectivity patterns. Our findings urge caution in adopting complex deep learning models for functional brain connectome analysis, emphasizing the need for rigorous experimental designs to establish tangible performance gains and perhaps more importantly, to pursue improvements in model interpretability.
ION-C: Integration of Overlapping Networks via Constraints
Nov 06, 2024Abstract:In many causal learning problems, variables of interest are often not all measured over the same observations, but are instead distributed across multiple datasets with overlapping variables. Tillman et al. (2008) presented the first algorithm for enumerating the minimal equivalence class of ground-truth DAGs consistent with all input graphs by exploiting local independence relations, called ION. In this paper, this problem is formulated as a more computationally efficient answer set programming (ASP) problem, which we call ION-C, and solved with the ASP system clingo. The ION-C algorithm was run on random synthetic graphs with varying sizes, densities, and degrees of overlap between subgraphs, with overlap having the largest impact on runtime, number of solution graphs, and agreement within the output set. To validate ION-C on real-world data, we ran the algorithm on overlapping graphs learned from data from two successive iterations of the European Social Survey (ESS), using a procedure for conducting joint independence tests to prevent inconsistencies in the input.
Unmasking Efficiency: Learning Salient Sparse Models in Non-IID Federated Learning
May 15, 2024



Abstract:In this work, we propose Salient Sparse Federated Learning (SSFL), a streamlined approach for sparse federated learning with efficient communication. SSFL identifies a sparse subnetwork prior to training, leveraging parameter saliency scores computed separately on local client data in non-IID scenarios, and then aggregated, to determine a global mask. Only the sparse model weights are communicated each round between the clients and the server. We validate SSFL's effectiveness using standard non-IID benchmarks, noting marked improvements in the sparsity--accuracy trade-offs. Finally, we deploy our method in a real-world federated learning framework and report improvement in communication time.
DynaLay: An Introspective Approach to Dynamic Layer Selection for Deep Networks
Dec 20, 2023Abstract:Deep learning models have become increasingly computationally intensive, requiring extensive computational resources and time for both training and inference. A significant contributing factor to this challenge is the uniform computational effort expended on each input example, regardless of its complexity. We introduce \textbf{DynaLay}, an alternative architecture that features a decision-making agent to adaptively select the most suitable layers for processing each input, thereby endowing the model with a remarkable level of introspection. DynaLay reevaluates more complex inputs during inference, adjusting the computational effort to optimize both performance and efficiency. The core of the system is a main model equipped with Fixed-Point Iterative (FPI) layers, capable of accurately approximating complex functions, paired with an agent that chooses these layers or a direct action based on the introspection of the models inner state. The model invests more time in processing harder examples, while minimal computation is required for easier ones. This introspective approach is a step toward developing deep learning models that "think" and "ponder", rather than "ballistically'' produce answers. Our experiments demonstrate that DynaLay achieves accuracy comparable to conventional deep models while significantly reducing computational demands.
Brainchop: Next Generation Web-Based Neuroimaging Application
Oct 24, 2023



Abstract:Performing volumetric image processing directly within the browser, particularly with medical data, presents unprecedented challenges compared to conventional backend tools. These challenges arise from limitations inherent in browser environments, such as constrained computational resources and the availability of frontend machine learning libraries. Consequently, there is a shortage of neuroimaging frontend tools capable of providing comprehensive end-to-end solutions for whole brain preprocessing and segmentation while preserving end-user data privacy and residency. In light of this context, we introduce Brainchop (http://www.brainchop.org) as a groundbreaking in-browser neuroimaging tool that enables volumetric analysis of structural MRI using pre-trained full-brain deep learning models, all without requiring technical expertise or intricate setup procedures. Beyond its commitment to data privacy, this frontend tool offers multiple features, including scalability, low latency, user-friendly operation, cross-platform compatibility, and enhanced accessibility. This paper outlines the processing pipeline of Brainchop and evaluates the performance of models across various software and hardware configurations. The results demonstrate the practicality of client-side processing for volumetric data, owing to the robust MeshNet architecture, even within the resource-constrained environment of web browsers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge