Lifang He
for the Alzheimer's Disease Neuroimaging Initiative
Digital Twin AI: Opportunities and Challenges from Large Language Models to World Models
Jan 04, 2026Abstract:Digital twins, as precise digital representations of physical systems, have evolved from passive simulation tools into intelligent and autonomous entities through the integration of artificial intelligence technologies. This paper presents a unified four-stage framework that systematically characterizes AI integration across the digital twin lifecycle, spanning modeling, mirroring, intervention, and autonomous management. By synthesizing existing technologies and practices, we distill a unified four-stage framework that systematically characterizes how AI methodologies are embedded across the digital twin lifecycle: (1) modeling the physical twin through physics-based and physics-informed AI approaches, (2) mirroring the physical system into a digital twin with real-time synchronization, (3) intervening in the physical twin through predictive modeling, anomaly detection, and optimization strategies, and (4) achieving autonomous management through large language models, foundation models, and intelligent agents. We analyze the synergy between physics-based modeling and data-driven learning, highlighting the shift from traditional numerical solvers to physics-informed and foundation models for physical systems. Furthermore, we examine how generative AI technologies, including large language models and generative world models, transform digital twins into proactive and self-improving cognitive systems capable of reasoning, communication, and creative scenario generation. Through a cross-domain review spanning eleven application domains, including healthcare, aerospace, smart manufacturing, robotics, and smart cities, we identify common challenges related to scalability, explainability, and trustworthiness, and outline directions for responsible AI-driven digital twin systems.
R-GenIMA: Integrating Neuroimaging and Genetics with Interpretable Multimodal AI for Alzheimer's Disease Progression
Dec 22, 2025Abstract:Early detection of Alzheimer's disease (AD) requires models capable of integrating macro-scale neuroanatomical alterations with micro-scale genetic susceptibility, yet existing multimodal approaches struggle to align these heterogeneous signals. We introduce R-GenIMA, an interpretable multimodal large language model that couples a novel ROI-wise vision transformer with genetic prompting to jointly model structural MRI and single nucleotide polymorphisms (SNPs) variations. By representing each anatomically parcellated brain region as a visual token and encoding SNP profiles as structured text, the framework enables cross-modal attention that links regional atrophy patterns to underlying genetic factors. Applied to the ADNI cohort, R-GenIMA achieves state-of-the-art performance in four-way classification across normal cognition (NC), subjective memory concerns (SMC), mild cognitive impairment (MCI), and AD. Beyond predictive accuracy, the model yields biologically meaningful explanations by identifying stage-specific brain regions and gene signatures, as well as coherent ROI-Gene association patterns across the disease continuum. Attention-based attribution revealed genes consistently enriched for established GWAS-supported AD risk loci, including APOE, BIN1, CLU, and RBFOX1. Stage-resolved neuroanatomical signatures identified shared vulnerability hubs across disease stages alongside stage-specific patterns: striatal involvement in subjective decline, frontotemporal engagement during prodromal impairment, and consolidated multimodal network disruption in AD. These results demonstrate that interpretable multimodal AI can synthesize imaging and genetics to reveal mechanistic insights, providing a foundation for clinically deployable tools that enable earlier risk stratification and inform precision therapeutic strategies in Alzheimer's disease.
Conditional Neural ODE for Longitudinal Parkinson's Disease Progression Forecasting
Nov 06, 2025Abstract:Parkinson's disease (PD) shows heterogeneous, evolving brain-morphometry patterns. Modeling these longitudinal trajectories enables mechanistic insight, treatment development, and individualized 'digital-twin' forecasting. However, existing methods usually adopt recurrent neural networks and transformer architectures, which rely on discrete, regularly sampled data while struggling to handle irregular and sparse magnetic resonance imaging (MRI) in PD cohorts. Moreover, these methods have difficulty capturing individual heterogeneity including variations in disease onset, progression rate, and symptom severity, which is a hallmark of PD. To address these challenges, we propose CNODE (Conditional Neural ODE), a novel framework for continuous, individualized PD progression forecasting. The core of CNODE is to model morphological brain changes as continuous temporal processes using a neural ODE model. In addition, we jointly learn patient-specific initial time and progress speed to align individual trajectories into a shared progression trajectory. We validate CNODE on the Parkinson's Progression Markers Initiative (PPMI) dataset. Experimental results show that our method outperforms state-of-the-art baselines in forecasting longitudinal PD progression.
Towards a general-purpose foundation model for fMRI analysis
Jun 11, 2025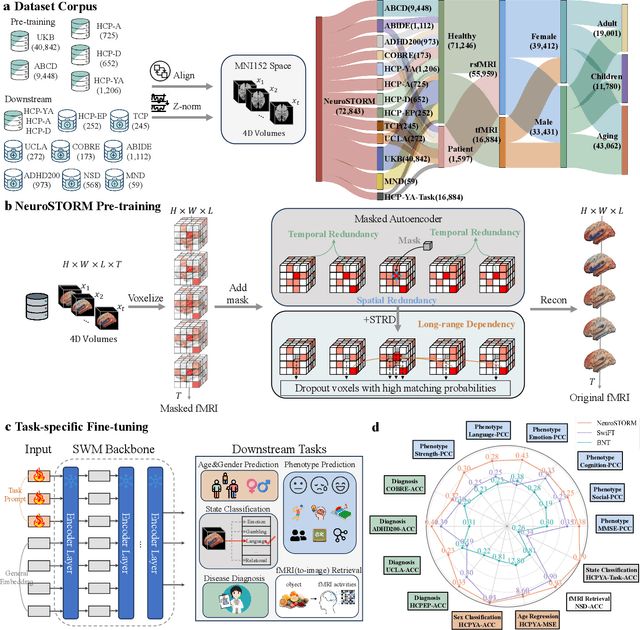

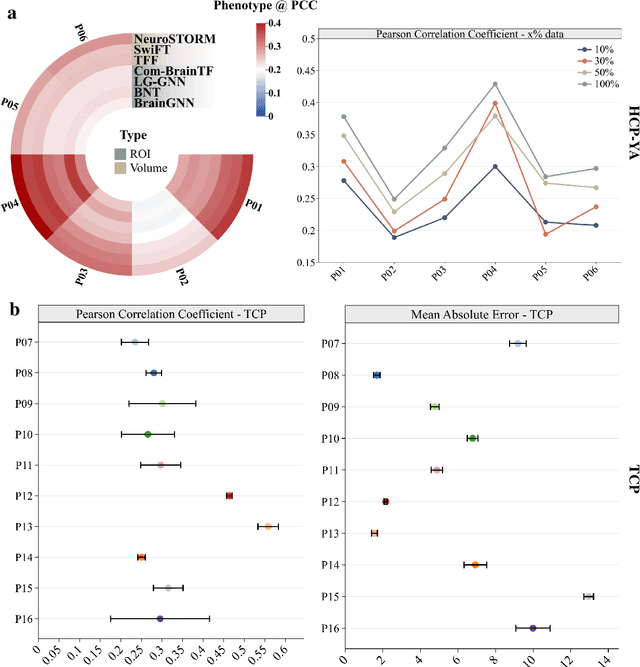
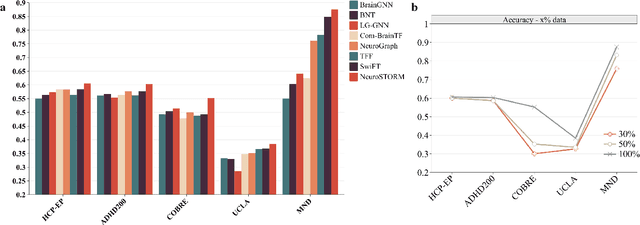
Abstract:Functional Magnetic Resonance Imaging (fMRI) is essential for studying brain function and diagnosing neurological disorders, but current analysis methods face reproducibility and transferability issues due to complex pre-processing and task-specific models. We introduce NeuroSTORM (Neuroimaging Foundation Model with Spatial-Temporal Optimized Representation Modeling), a generalizable framework that directly learns from 4D fMRI volumes and enables efficient knowledge transfer across diverse applications. NeuroSTORM is pre-trained on 28.65 million fMRI frames (>9,000 hours) from over 50,000 subjects across multiple centers and ages 5 to 100. Using a Mamba backbone and a shifted scanning strategy, it efficiently processes full 4D volumes. We also propose a spatial-temporal optimized pre-training approach and task-specific prompt tuning to improve transferability. NeuroSTORM outperforms existing methods across five tasks: age/gender prediction, phenotype prediction, disease diagnosis, fMRI-to-image retrieval, and task-based fMRI classification. It demonstrates strong clinical utility on datasets from hospitals in the U.S., South Korea, and Australia, achieving top performance in disease diagnosis and cognitive phenotype prediction. NeuroSTORM provides a standardized, open-source foundation model to improve reproducibility and transferability in fMRI-based clinical research.
EfficientLLM: Efficiency in Large Language Models
May 20, 2025Abstract:Large Language Models (LLMs) have driven significant progress, yet their growing parameter counts and context windows incur prohibitive compute, energy, and monetary costs. We introduce EfficientLLM, a novel benchmark and the first comprehensive empirical study evaluating efficiency techniques for LLMs at scale. Conducted on a production-class cluster (48xGH200, 8xH200 GPUs), our study systematically explores three key axes: (1) architecture pretraining (efficient attention variants: MQA, GQA, MLA, NSA; sparse Mixture-of-Experts (MoE)), (2) fine-tuning (parameter-efficient methods: LoRA, RSLoRA, DoRA), and (3) inference (quantization methods: int4, float16). We define six fine-grained metrics (Memory Utilization, Compute Utilization, Latency, Throughput, Energy Consumption, Compression Rate) to capture hardware saturation, latency-throughput balance, and carbon cost. Evaluating over 100 model-technique pairs (0.5B-72B parameters), we derive three core insights: (i) Efficiency involves quantifiable trade-offs: no single method is universally optimal; e.g., MoE reduces FLOPs and improves accuracy but increases VRAM by 40%, while int4 quantization cuts memory/energy by up to 3.9x at a 3-5% accuracy drop. (ii) Optima are task- and scale-dependent: MQA offers optimal memory-latency trade-offs for constrained devices, MLA achieves lowest perplexity for quality-critical tasks, and RSLoRA surpasses LoRA efficiency only beyond 14B parameters. (iii) Techniques generalize across modalities: we extend evaluations to Large Vision Models (Stable Diffusion 3.5, Wan 2.1) and Vision-Language Models (Qwen2.5-VL), confirming effective transferability. By open-sourcing datasets, evaluation pipelines, and leaderboards, EfficientLLM provides essential guidance for researchers and engineers navigating the efficiency-performance landscape of next-generation foundation models.
A Survey on Post-training of Large Language Models
Mar 08, 2025Abstract:The emergence of Large Language Models (LLMs) has fundamentally transformed natural language processing, making them indispensable across domains ranging from conversational systems to scientific exploration. However, their pre-trained architectures often reveal limitations in specialized contexts, including restricted reasoning capacities, ethical uncertainties, and suboptimal domain-specific performance. These challenges necessitate advanced post-training language models (PoLMs) to address these shortcomings, such as OpenAI-o1/o3 and DeepSeek-R1 (collectively known as Large Reasoning Models, or LRMs). This paper presents the first comprehensive survey of PoLMs, systematically tracing their evolution across five core paradigms: Fine-tuning, which enhances task-specific accuracy; Alignment, which ensures alignment with human preferences; Reasoning, which advances multi-step inference despite challenges in reward design; Efficiency, which optimizes resource utilization amidst increasing complexity; and Integration and Adaptation, which extend capabilities across diverse modalities while addressing coherence issues. Charting progress from ChatGPT's foundational alignment strategies to DeepSeek-R1's innovative reasoning advancements, we illustrate how PoLMs leverage datasets to mitigate biases, deepen reasoning capabilities, and enhance domain adaptability. Our contributions include a pioneering synthesis of PoLM evolution, a structured taxonomy categorizing techniques and datasets, and a strategic agenda emphasizing the role of LRMs in improving reasoning proficiency and domain flexibility. As the first survey of its scope, this work consolidates recent PoLM advancements and establishes a rigorous intellectual framework for future research, fostering the development of LLMs that excel in precision, ethical robustness, and versatility across scientific and societal applications.
End-to-End Deep Learning for Structural Brain Imaging: A Unified Framework
Feb 23, 2025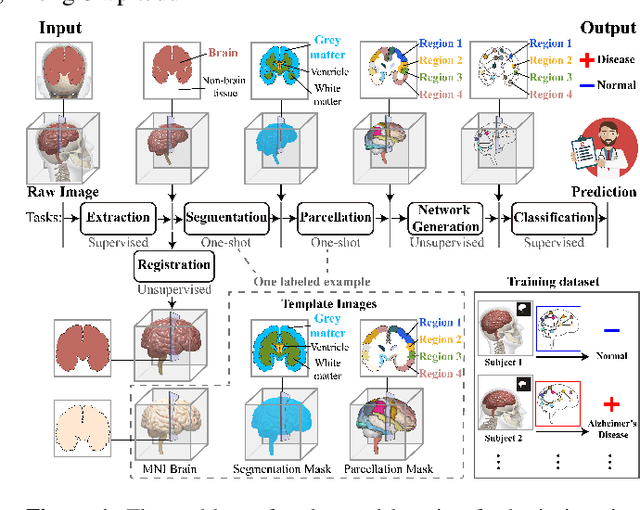
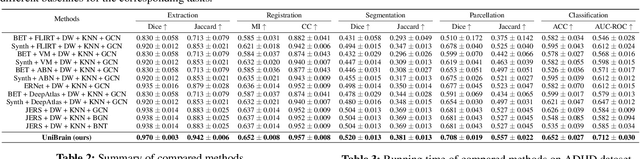
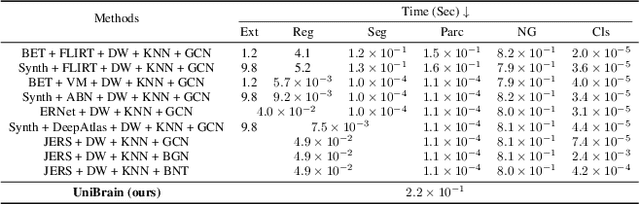
Abstract:Brain imaging analysis is fundamental in neuroscience, providing valuable insights into brain structure and function. Traditional workflows follow a sequential pipeline-brain extraction, registration, segmentation, parcellation, network generation, and classification-treating each step as an independent task. These methods rely heavily on task-specific training data and expert intervention to correct intermediate errors, making them particularly burdensome for high-dimensional neuroimaging data, where annotations and quality control are costly and time-consuming. We introduce UniBrain, a unified end-to-end framework that integrates all processing steps into a single optimization process, allowing tasks to interact and refine each other. Unlike traditional approaches that require extensive task-specific annotations, UniBrain operates with minimal supervision, leveraging only low-cost labels (i.e., classification and extraction) and a single labeled atlas. By jointly optimizing extraction, registration, segmentation, parcellation, network generation, and classification, UniBrain enhances both accuracy and computational efficiency while significantly reducing annotation effort. Experimental results demonstrate its superiority over existing methods across multiple tasks, offering a more scalable and reliable solution for neuroimaging analysis. Our code and data can be found at https://github.com/Anonymous7852/UniBrain
Rethinking Functional Brain Connectome Analysis: Do Graph Deep Learning Models Help?
Jan 28, 2025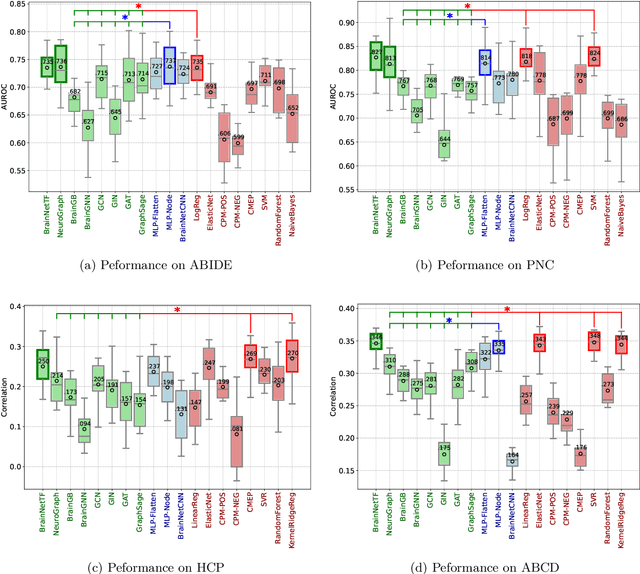
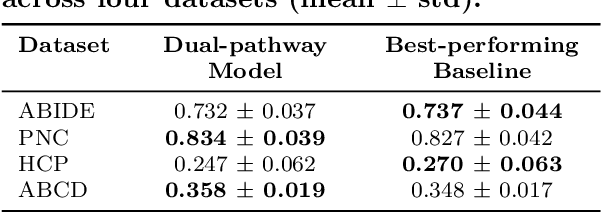
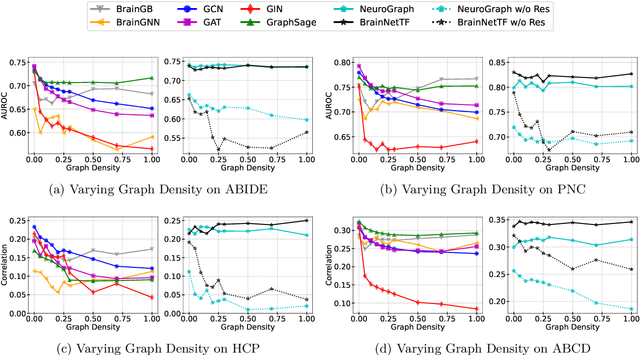
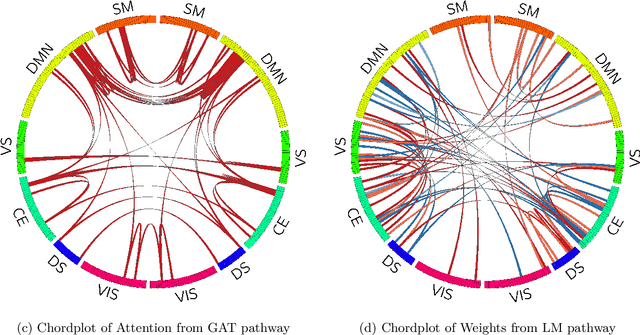
Abstract:Functional brain connectome is crucial for deciphering the neural mechanisms underlying cognitive functions and neurological disorders. Graph deep learning models have recently gained tremendous popularity in this field. However, their actual effectiveness in modeling the brain connectome remains unclear. In this study, we re-examine graph deep learning models based on four large-scale neuroimaging studies encompassing diverse cognitive and clinical outcomes. Surprisingly, we find that the message aggregation mechanism, a hallmark of graph deep learning models, does not help with predictive performance as typically assumed, but rather consistently degrades it. To address this issue, we propose a hybrid model combining a linear model with a graph attention network through dual pathways, achieving robust predictions and enhanced interpretability by revealing both localized and global neural connectivity patterns. Our findings urge caution in adopting complex deep learning models for functional brain connectome analysis, emphasizing the need for rigorous experimental designs to establish tangible performance gains and perhaps more importantly, to pursue improvements in model interpretability.
Large Language Models for Bioinformatics
Jan 10, 2025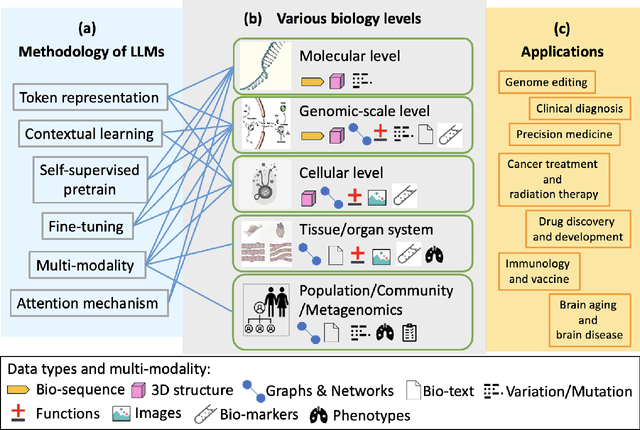
Abstract:With the rapid advancements in large language model (LLM) technology and the emergence of bioinformatics-specific language models (BioLMs), there is a growing need for a comprehensive analysis of the current landscape, computational characteristics, and diverse applications. This survey aims to address this need by providing a thorough review of BioLMs, focusing on their evolution, classification, and distinguishing features, alongside a detailed examination of training methodologies, datasets, and evaluation frameworks. We explore the wide-ranging applications of BioLMs in critical areas such as disease diagnosis, drug discovery, and vaccine development, highlighting their impact and transformative potential in bioinformatics. We identify key challenges and limitations inherent in BioLMs, including data privacy and security concerns, interpretability issues, biases in training data and model outputs, and domain adaptation complexities. Finally, we highlight emerging trends and future directions, offering valuable insights to guide researchers and clinicians toward advancing BioLMs for increasingly sophisticated biological and clinical applications.
Political-LLM: Large Language Models in Political Science
Dec 09, 2024



Abstract:In recent years, large language models (LLMs) have been widely adopted in political science tasks such as election prediction, sentiment analysis, policy impact assessment, and misinformation detection. Meanwhile, the need to systematically understand how LLMs can further revolutionize the field also becomes urgent. In this work, we--a multidisciplinary team of researchers spanning computer science and political science--present the first principled framework termed Political-LLM to advance the comprehensive understanding of integrating LLMs into computational political science. Specifically, we first introduce a fundamental taxonomy classifying the existing explorations into two perspectives: political science and computational methodologies. In particular, from the political science perspective, we highlight the role of LLMs in automating predictive and generative tasks, simulating behavior dynamics, and improving causal inference through tools like counterfactual generation; from a computational perspective, we introduce advancements in data preparation, fine-tuning, and evaluation methods for LLMs that are tailored to political contexts. We identify key challenges and future directions, emphasizing the development of domain-specific datasets, addressing issues of bias and fairness, incorporating human expertise, and redefining evaluation criteria to align with the unique requirements of computational political science. Political-LLM seeks to serve as a guidebook for researchers to foster an informed, ethical, and impactful use of Artificial Intelligence in political science. Our online resource is available at: http://political-llm.org/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge