Rong Zhou
Digital Twin AI: Opportunities and Challenges from Large Language Models to World Models
Jan 04, 2026Abstract:Digital twins, as precise digital representations of physical systems, have evolved from passive simulation tools into intelligent and autonomous entities through the integration of artificial intelligence technologies. This paper presents a unified four-stage framework that systematically characterizes AI integration across the digital twin lifecycle, spanning modeling, mirroring, intervention, and autonomous management. By synthesizing existing technologies and practices, we distill a unified four-stage framework that systematically characterizes how AI methodologies are embedded across the digital twin lifecycle: (1) modeling the physical twin through physics-based and physics-informed AI approaches, (2) mirroring the physical system into a digital twin with real-time synchronization, (3) intervening in the physical twin through predictive modeling, anomaly detection, and optimization strategies, and (4) achieving autonomous management through large language models, foundation models, and intelligent agents. We analyze the synergy between physics-based modeling and data-driven learning, highlighting the shift from traditional numerical solvers to physics-informed and foundation models for physical systems. Furthermore, we examine how generative AI technologies, including large language models and generative world models, transform digital twins into proactive and self-improving cognitive systems capable of reasoning, communication, and creative scenario generation. Through a cross-domain review spanning eleven application domains, including healthcare, aerospace, smart manufacturing, robotics, and smart cities, we identify common challenges related to scalability, explainability, and trustworthiness, and outline directions for responsible AI-driven digital twin systems.
EfficientLLM: Efficiency in Large Language Models
May 20, 2025Abstract:Large Language Models (LLMs) have driven significant progress, yet their growing parameter counts and context windows incur prohibitive compute, energy, and monetary costs. We introduce EfficientLLM, a novel benchmark and the first comprehensive empirical study evaluating efficiency techniques for LLMs at scale. Conducted on a production-class cluster (48xGH200, 8xH200 GPUs), our study systematically explores three key axes: (1) architecture pretraining (efficient attention variants: MQA, GQA, MLA, NSA; sparse Mixture-of-Experts (MoE)), (2) fine-tuning (parameter-efficient methods: LoRA, RSLoRA, DoRA), and (3) inference (quantization methods: int4, float16). We define six fine-grained metrics (Memory Utilization, Compute Utilization, Latency, Throughput, Energy Consumption, Compression Rate) to capture hardware saturation, latency-throughput balance, and carbon cost. Evaluating over 100 model-technique pairs (0.5B-72B parameters), we derive three core insights: (i) Efficiency involves quantifiable trade-offs: no single method is universally optimal; e.g., MoE reduces FLOPs and improves accuracy but increases VRAM by 40%, while int4 quantization cuts memory/energy by up to 3.9x at a 3-5% accuracy drop. (ii) Optima are task- and scale-dependent: MQA offers optimal memory-latency trade-offs for constrained devices, MLA achieves lowest perplexity for quality-critical tasks, and RSLoRA surpasses LoRA efficiency only beyond 14B parameters. (iii) Techniques generalize across modalities: we extend evaluations to Large Vision Models (Stable Diffusion 3.5, Wan 2.1) and Vision-Language Models (Qwen2.5-VL), confirming effective transferability. By open-sourcing datasets, evaluation pipelines, and leaderboards, EfficientLLM provides essential guidance for researchers and engineers navigating the efficiency-performance landscape of next-generation foundation models.
ELITE: Embedding-Less retrieval with Iterative Text Exploration
May 17, 2025Abstract:Large Language Models (LLMs) have achieved impressive progress in natural language processing, but their limited ability to retain long-term context constrains performance on document-level or multi-turn tasks. Retrieval-Augmented Generation (RAG) mitigates this by retrieving relevant information from an external corpus. However, existing RAG systems often rely on embedding-based retrieval trained on corpus-level semantic similarity, which can lead to retrieving content that is semantically similar in form but misaligned with the question's true intent. Furthermore, recent RAG variants construct graph- or hierarchy-based structures to improve retrieval accuracy, resulting in significant computation and storage overhead. In this paper, we propose an embedding-free retrieval framework. Our method leverages the logical inferencing ability of LLMs in retrieval using iterative search space refinement guided by our novel importance measure and extend our retrieval results with logically related information without explicit graph construction. Experiments on long-context QA benchmarks, including NovelQA and Marathon, show that our approach outperforms strong baselines while reducing storage and runtime by over an order of magnitude.
The study of non-complete-ring positron emission tomography (PET) detection method
Apr 01, 2025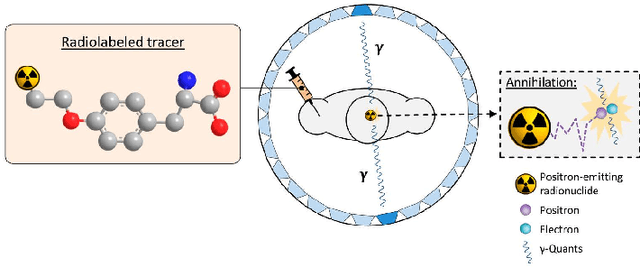
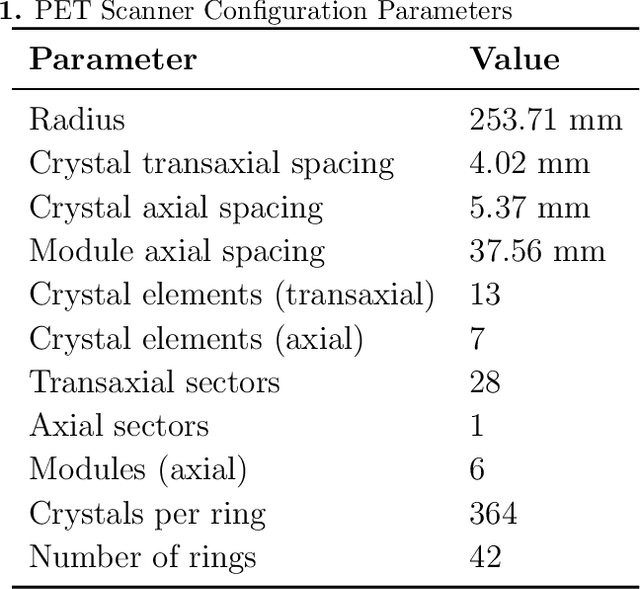
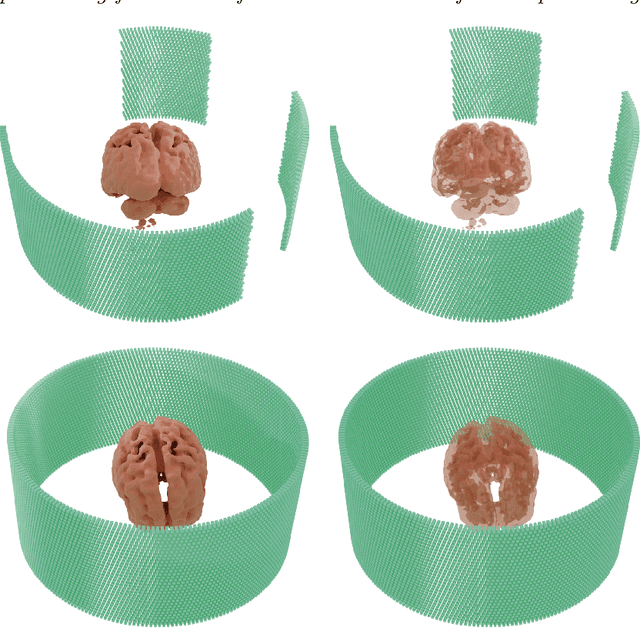
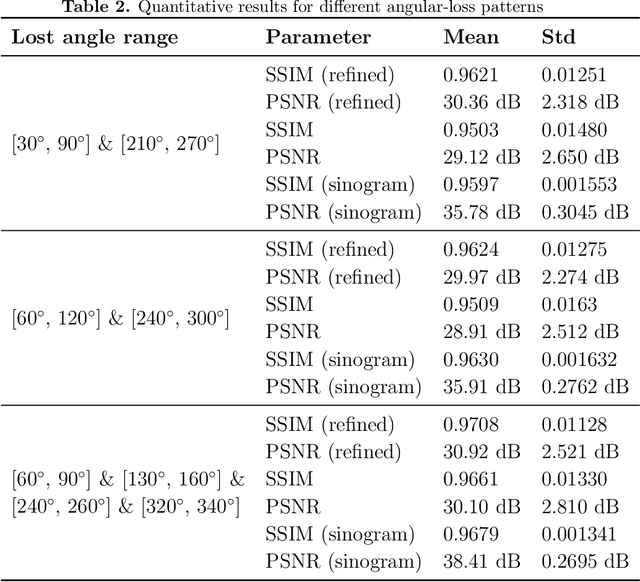
Abstract:Positron Emission Tomography (PET) is a vital molecular imaging tool widely used in medical diagnosis and treatment evaluation. Traditional PET systems typically rely on complete detector rings to achieve full angular coverage for uniform and statistically robust sampling of coincidence events. However, incomplete-ring PET scanners have emerged in various scenarios due to hardware failures, cost constraints, or specific clinical needs. In such cases, conventional reconstruction algorithms often suffer from performance degradation due to reduced data completeness and geometric inconsistencies. This thesis proposes a coarse-to-fine reconstruction framework for incomplete-ring PET scanners. The framework first employs an Attention U-Net model to recover complete sinograms from incomplete ones, then uses the OSEM algorithm for preliminary reconstruction, and finally applies a two-stage architecture comprising a Coarse Prediction Module (CPM) and an Iterative Refinement Module (IRM) for fine reconstruction. Our approach utilizes neighboring axial slices and spectral transform features as auxiliary guidance at the input level to ensure spatial and frequency domain consistency, and integrates a contrastive diffusion strategy at the output level to improve correspondence between low-quality PET inputs and refined PET outputs. Experimental results on public and in-house brain PET datasets demonstrate that the proposed method significantly outperforms existing approaches in metrics such as PSNR (35.6421 dB) and SSIM (0.9588), successfully preserving key anatomical structures and tracer distribution features, thus providing an effective solution for incomplete-ring PET imaging.
A Survey on Post-training of Large Language Models
Mar 08, 2025Abstract:The emergence of Large Language Models (LLMs) has fundamentally transformed natural language processing, making them indispensable across domains ranging from conversational systems to scientific exploration. However, their pre-trained architectures often reveal limitations in specialized contexts, including restricted reasoning capacities, ethical uncertainties, and suboptimal domain-specific performance. These challenges necessitate advanced post-training language models (PoLMs) to address these shortcomings, such as OpenAI-o1/o3 and DeepSeek-R1 (collectively known as Large Reasoning Models, or LRMs). This paper presents the first comprehensive survey of PoLMs, systematically tracing their evolution across five core paradigms: Fine-tuning, which enhances task-specific accuracy; Alignment, which ensures alignment with human preferences; Reasoning, which advances multi-step inference despite challenges in reward design; Efficiency, which optimizes resource utilization amidst increasing complexity; and Integration and Adaptation, which extend capabilities across diverse modalities while addressing coherence issues. Charting progress from ChatGPT's foundational alignment strategies to DeepSeek-R1's innovative reasoning advancements, we illustrate how PoLMs leverage datasets to mitigate biases, deepen reasoning capabilities, and enhance domain adaptability. Our contributions include a pioneering synthesis of PoLM evolution, a structured taxonomy categorizing techniques and datasets, and a strategic agenda emphasizing the role of LRMs in improving reasoning proficiency and domain flexibility. As the first survey of its scope, this work consolidates recent PoLM advancements and establishes a rigorous intellectual framework for future research, fostering the development of LLMs that excel in precision, ethical robustness, and versatility across scientific and societal applications.
Normative Modeling for AD Diagnosis and Biomarker Identification
Nov 15, 2024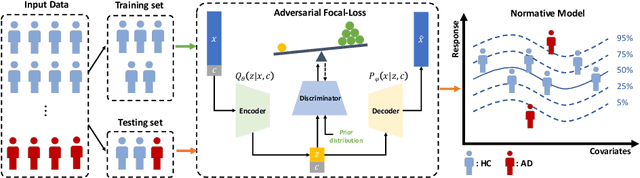
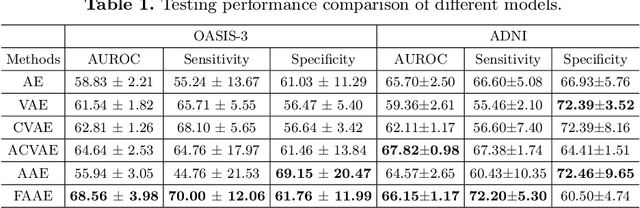
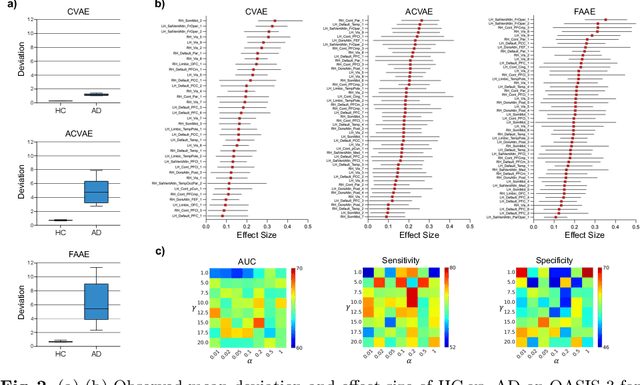
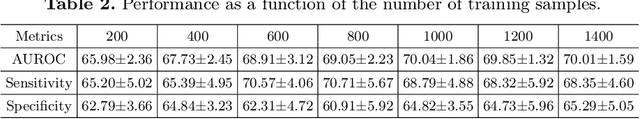
Abstract:In this paper, we introduce a novel normative modeling approach that incorporates focal loss and adversarial autoencoders (FAAE) for Alzheimer's Disease (AD) diagnosis and biomarker identification. Our method is an end-to-end approach that embeds an adversarial focal loss discriminator within the autoencoder structure, specifically designed to effectively target and capture more complex and challenging cases. We first use the enhanced autoencoder to create a normative model based on data from healthy control (HC) individuals. We then apply this model to estimate total and regional neuroanatomical deviation in AD patients. Through extensive experiments on the OASIS-3 and ADNI datasets, our approach significantly outperforms previous state-of-the-art methods. This advancement not only streamlines the detection process but also provides a greater insight into the biomarker potential for AD. Our code can be found at \url{https://github.com/soz223/FAAE}.
TTT-Unet: Enhancing U-Net with Test-Time Training Layers for Biomedical Image Segmentation
Sep 18, 2024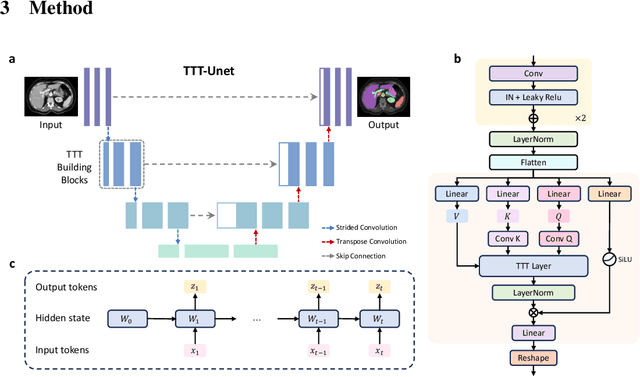

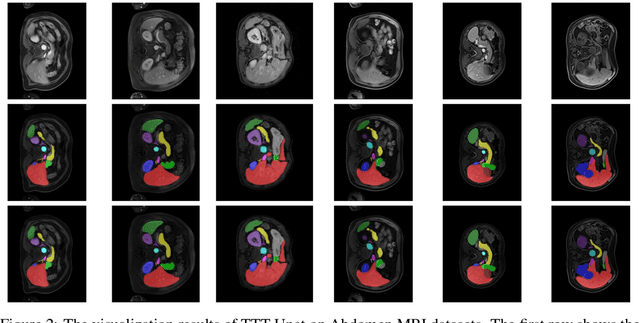
Abstract:Biomedical image segmentation is crucial for accurately diagnosing and analyzing various diseases. However, Convolutional Neural Networks (CNNs) and Transformers, the most commonly used architectures for this task, struggle to effectively capture long-range dependencies due to the inherent locality of CNNs and the computational complexity of Transformers. To address this limitation, we introduce TTT-Unet, a novel framework that integrates Test-Time Training (TTT) layers into the traditional U-Net architecture for biomedical image segmentation. TTT-Unet dynamically adjusts model parameters during the testing time, enhancing the model's ability to capture both local and long-range features. We evaluate TTT-Unet on multiple medical imaging datasets, including 3D abdominal organ segmentation in CT and MR images, instrument segmentation in endoscopy images, and cell segmentation in microscopy images. The results demonstrate that TTT-Unet consistently outperforms state-of-the-art CNN-based and Transformer-based segmentation models across all tasks. The code is available at https://github.com/rongzhou7/TTT-Unet.
Biomedical SAM 2: Segment Anything in Biomedical Images and Videos
Aug 06, 2024
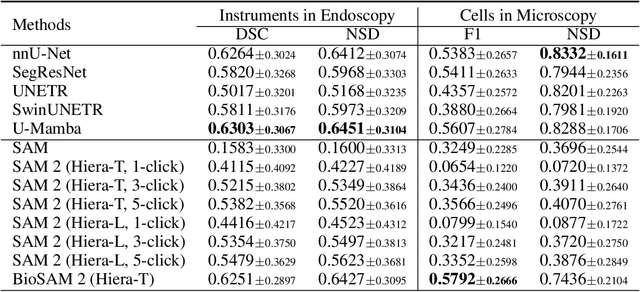
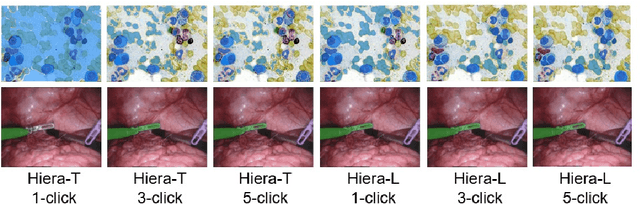
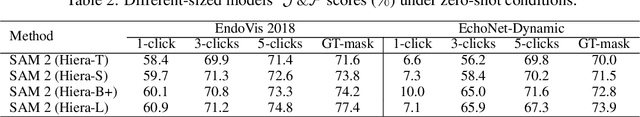
Abstract:Medical image segmentation and video object segmentation are essential for diagnosing and analyzing diseases by identifying and measuring biological structures. Recent advances in natural domain have been driven by foundation models like the Segment Anything Model 2 (SAM 2). To explore the performance of SAM 2 in biomedical applications, we designed two evaluation pipelines for single-frame image segmentation and multi-frame video segmentation with varied prompt designs, revealing SAM 2's limitations in medical contexts. Consequently, we developed BioSAM 2, an enhanced foundation model optimized for biomedical data based on SAM 2. Our experiments show that BioSAM 2 not only surpasses the performance of existing state-of-the-art foundation models but also matches or even exceeds specialist models, demonstrating its efficacy and potential in the medical domain.
ViT-1.58b: Mobile Vision Transformers in the 1-bit Era
Jun 26, 2024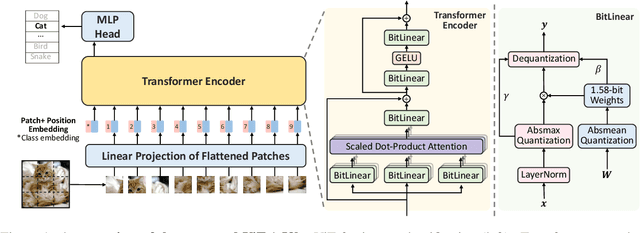
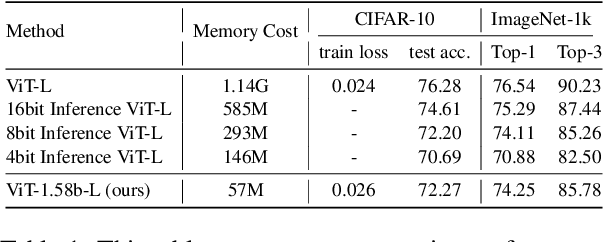
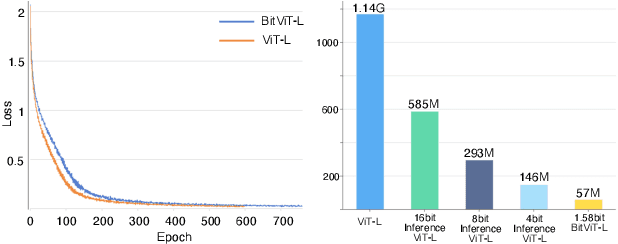
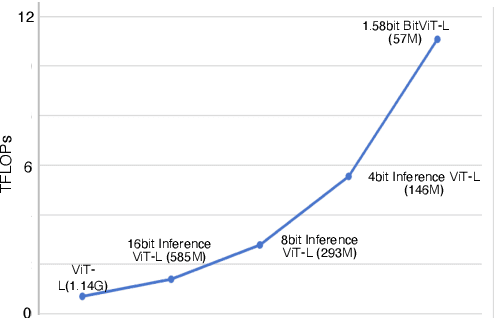
Abstract:Vision Transformers (ViTs) have achieved remarkable performance in various image classification tasks by leveraging the attention mechanism to process image patches as tokens. However, the high computational and memory demands of ViTs pose significant challenges for deployment in resource-constrained environments. This paper introduces ViT-1.58b, a novel 1.58-bit quantized ViT model designed to drastically reduce memory and computational overhead while preserving competitive performance. ViT-1.58b employs ternary quantization, which refines the balance between efficiency and accuracy by constraining weights to {-1, 0, 1} and quantizing activations to 8-bit precision. Our approach ensures efficient scaling in terms of both memory and computation. Experiments on CIFAR-10 and ImageNet-1k demonstrate that ViT-1.58b maintains comparable accuracy to full-precision Vit, with significant reductions in memory usage and computational costs. This paper highlights the potential of extreme quantization techniques in developing sustainable AI solutions and contributes to the broader discourse on efficient model deployment in practical applications. Our code and weights are available at https://github.com/DLYuanGod/ViT-1.58b.
Unleashing the Power of Multi-Task Learning: A Comprehensive Survey Spanning Traditional, Deep, and Pretrained Foundation Model Eras
Apr 29, 2024



Abstract:MTL is a learning paradigm that effectively leverages both task-specific and shared information to address multiple related tasks simultaneously. In contrast to STL, MTL offers a suite of benefits that enhance both the training process and the inference efficiency. MTL's key advantages encompass streamlined model architecture, performance enhancement, and cross-domain generalizability. Over the past twenty years, MTL has become widely recognized as a flexible and effective approach in various fields, including CV, NLP, recommendation systems, disease prognosis and diagnosis, and robotics. This survey provides a comprehensive overview of the evolution of MTL, encompassing the technical aspects of cutting-edge methods from traditional approaches to deep learning and the latest trend of pretrained foundation models. Our survey methodically categorizes MTL techniques into five key areas: regularization, relationship learning, feature propagation, optimization, and pre-training. This categorization not only chronologically outlines the development of MTL but also dives into various specialized strategies within each category. Furthermore, the survey reveals how the MTL evolves from handling a fixed set of tasks to embracing a more flexible approach free from task or modality constraints. It explores the concepts of task-promptable and -agnostic training, along with the capacity for ZSL, which unleashes the untapped potential of this historically coveted learning paradigm. Overall, we hope this survey provides the research community with a comprehensive overview of the advancements in MTL from its inception in 1997 to the present in 2023. We address present challenges and look ahead to future possibilities, shedding light on the opportunities and potential avenues for MTL research in a broad manner. This project is publicly available at https://github.com/junfish/Awesome-Multitask-Learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge