Yiwei Li
Learning More from Less: Unlocking Internal Representations for Benchmark Compression
Feb 03, 2026Abstract:The prohibitive cost of evaluating Large Language Models (LLMs) necessitates efficient alternatives to full-scale benchmarking. Prevalent approaches address this by identifying a small coreset of items to approximate full-benchmark performance. However, existing methods must estimate a reliable item profile from response patterns across many source models, which becomes statistically unstable when the source pool is small. This dependency is particularly limiting for newly released benchmarks with minimal historical evaluation data. We argue that discrete correctness labels are a lossy view of the model's decision process and fail to capture information encoded in hidden states. To address this, we introduce REPCORE, which aligns heterogeneous hidden states into a unified latent space to construct representative coresets. Using these subsets for performance extrapolation, REPCORE achieves precise estimation accuracy with as few as ten source models. Experiments on five benchmarks and over 200 models show consistent gains over output-based baselines in ranking correlation and estimation accuracy. Spectral analysis further indicates that the aligned representations contain separable components reflecting broad response tendencies and task-specific reasoning patterns.
Kimi K2.5: Visual Agentic Intelligence
Feb 02, 2026Abstract:We introduce Kimi K2.5, an open-source multimodal agentic model designed to advance general agentic intelligence. K2.5 emphasizes the joint optimization of text and vision so that two modalities enhance each other. This includes a series of techniques such as joint text-vision pre-training, zero-vision SFT, and joint text-vision reinforcement learning. Building on this multimodal foundation, K2.5 introduces Agent Swarm, a self-directed parallel agent orchestration framework that dynamically decomposes complex tasks into heterogeneous sub-problems and executes them concurrently. Extensive evaluations show that Kimi K2.5 achieves state-of-the-art results across various domains including coding, vision, reasoning, and agentic tasks. Agent Swarm also reduces latency by up to $4.5\times$ over single-agent baselines. We release the post-trained Kimi K2.5 model checkpoint to facilitate future research and real-world applications of agentic intelligence.
Do Not Waste Your Rollouts: Recycling Search Experience for Efficient Test-Time Scaling
Jan 29, 2026Abstract:Test-Time Scaling enhances the reasoning capabilities of Large Language Models by allocating additional inference compute to broaden the exploration of the solution space. However, existing search strategies typically treat rollouts as disposable samples, where valuable intermediate insights are effectively discarded after each trial. This systemic memorylessness leads to massive computational redundancy, as models repeatedly re-derive discovered conclusions and revisit known dead ends across extensive attempts. To bridge this gap, we propose \textbf{Recycling Search Experience (RSE)}, a self-guided, training-free strategy that turns test-time search from a series of isolated trials into a cumulative process. By actively distilling raw trajectories into a shared experience bank, RSE enables positive recycling of intermediate conclusions to shortcut redundant derivations and negative recycling of failure patterns to prune encountered dead ends. Theoretically, we provide an analysis that formalizes the efficiency gains of RSE, validating its advantage over independent sampling in solving complex reasoning tasks. Empirically, extensive experiments on HMMT24, HMMT25, IMO-Bench, and HLE show that RSE consistently outperforms strong baselines with comparable computational cost, achieving state-of-the-art scaling efficiency.
Kimi Linear: An Expressive, Efficient Attention Architecture
Oct 30, 2025Abstract:We introduce Kimi Linear, a hybrid linear attention architecture that, for the first time, outperforms full attention under fair comparisons across various scenarios -- including short-context, long-context, and reinforcement learning (RL) scaling regimes. At its core lies Kimi Delta Attention (KDA), an expressive linear attention module that extends Gated DeltaNet with a finer-grained gating mechanism, enabling more effective use of limited finite-state RNN memory. Our bespoke chunkwise algorithm achieves high hardware efficiency through a specialized variant of the Diagonal-Plus-Low-Rank (DPLR) transition matrices, which substantially reduces computation compared to the general DPLR formulation while remaining more consistent with the classical delta rule. We pretrain a Kimi Linear model with 3B activated parameters and 48B total parameters, based on a layerwise hybrid of KDA and Multi-Head Latent Attention (MLA). Our experiments show that with an identical training recipe, Kimi Linear outperforms full MLA with a sizeable margin across all evaluated tasks, while reducing KV cache usage by up to 75% and achieving up to 6 times decoding throughput for a 1M context. These results demonstrate that Kimi Linear can be a drop-in replacement for full attention architectures with superior performance and efficiency, including tasks with longer input and output lengths. To support further research, we open-source the KDA kernel and vLLM implementations, and release the pre-trained and instruction-tuned model checkpoints.
RAU: Reference-based Anatomical Understanding with Vision Language Models
Sep 26, 2025Abstract:Anatomical understanding through deep learning is critical for automatic report generation, intra-operative navigation, and organ localization in medical imaging; however, its progress is constrained by the scarcity of expert-labeled data. A promising remedy is to leverage an annotated reference image to guide the interpretation of an unlabeled target. Although recent vision-language models (VLMs) exhibit non-trivial visual reasoning, their reference-based understanding and fine-grained localization remain limited. We introduce RAU, a framework for reference-based anatomical understanding with VLMs. We first show that a VLM learns to identify anatomical regions through relative spatial reasoning between reference and target images, trained on a moderately sized dataset. We validate this capability through visual question answering (VQA) and bounding box prediction. Next, we demonstrate that the VLM-derived spatial cues can be seamlessly integrated with the fine-grained segmentation capability of SAM2, enabling localization and pixel-level segmentation of small anatomical regions, such as vessel segments. Across two in-distribution and two out-of-distribution datasets, RAU consistently outperforms a SAM2 fine-tuning baseline using the same memory setup, yielding more accurate segmentations and more reliable localization. More importantly, its strong generalization ability makes it scalable to out-of-distribution datasets, a property crucial for medical image applications. To the best of our knowledge, RAU is the first to explore the capability of VLMs for reference-based identification, localization, and segmentation of anatomical structures in medical images. Its promising performance highlights the potential of VLM-driven approaches for anatomical understanding in automated clinical workflows.
Memory Enhanced Fractional-Order Dung Beetle Optimization for Photovoltaic Parameter Identification
Aug 09, 2025Abstract:Accurate parameter identification in photovoltaic (PV) models is crucial for performance evaluation but remains challenging due to their nonlinear, multimodal, and high-dimensional nature. Although the Dung Beetle Optimization (DBO) algorithm has shown potential in addressing such problems, it often suffers from premature convergence. To overcome these issues, this paper proposes a Memory Enhanced Fractional-Order Dung Beetle Optimization (MFO-DBO) algorithm that integrates three coordinated strategies. Firstly, fractional-order (FO) calculus introduces memory into the search process, enhancing convergence stability and solution quality. Secondly, a fractional-order logistic chaotic map improves population diversity during initialization. Thirdly, a chaotic perturbation mechanism helps elite solutions escape local optima. Numerical results on the CEC2017 benchmark suite and the PV parameter identification problem demonstrate that MFO-DBO consistently outperforms advanced DBO variants, CEC competition winners, FO-based optimizers, enhanced classical algorithms, and recent metaheuristics in terms of accuracy, robustness, convergence speed, while also maintaining an excellent balance between exploration and exploitation compared to the standard DBO algorithm.
Kimi K2: Open Agentic Intelligence
Jul 28, 2025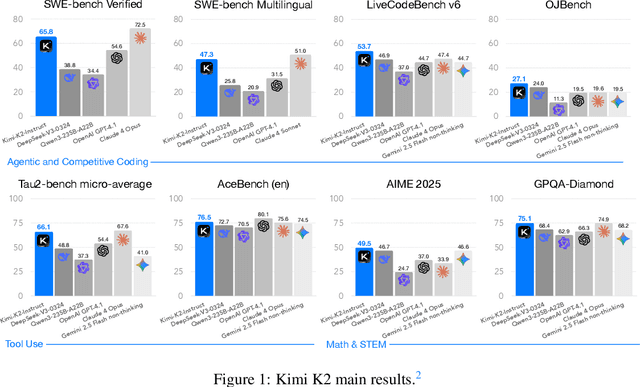

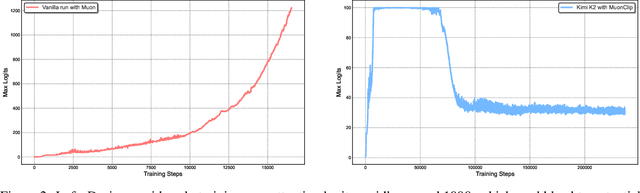
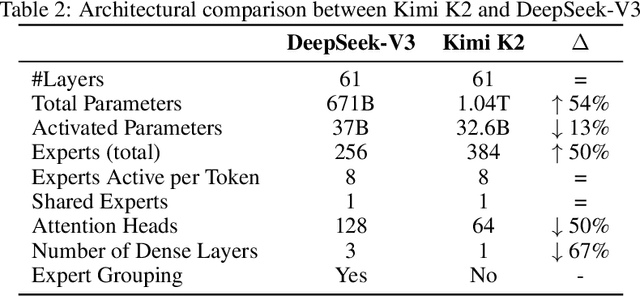
Abstract:We introduce Kimi K2, a Mixture-of-Experts (MoE) large language model with 32 billion activated parameters and 1 trillion total parameters. We propose the MuonClip optimizer, which improves upon Muon with a novel QK-clip technique to address training instability while enjoying the advanced token efficiency of Muon. Based on MuonClip, K2 was pre-trained on 15.5 trillion tokens with zero loss spike. During post-training, K2 undergoes a multi-stage post-training process, highlighted by a large-scale agentic data synthesis pipeline and a joint reinforcement learning (RL) stage, where the model improves its capabilities through interactions with real and synthetic environments. Kimi K2 achieves state-of-the-art performance among open-source non-thinking models, with strengths in agentic capabilities. Notably, K2 obtains 66.1 on Tau2-Bench, 76.5 on ACEBench (En), 65.8 on SWE-Bench Verified, and 47.3 on SWE-Bench Multilingual -- surpassing most open and closed-sourced baselines in non-thinking settings. It also exhibits strong capabilities in coding, mathematics, and reasoning tasks, with a score of 53.7 on LiveCodeBench v6, 49.5 on AIME 2025, 75.1 on GPQA-Diamond, and 27.1 on OJBench, all without extended thinking. These results position Kimi K2 as one of the most capable open-source large language models to date, particularly in software engineering and agentic tasks. We release our base and post-trained model checkpoints to facilitate future research and applications of agentic intelligence.
Mind the Quote: Enabling Quotation-Aware Dialogue in LLMs via Plug-and-Play Modules
May 30, 2025Abstract:Human-AI conversation frequently relies on quoting earlier text-"check it with the formula I just highlighted"-yet today's large language models (LLMs) lack an explicit mechanism for locating and exploiting such spans. We formalise the challenge as span-conditioned generation, decomposing each turn into the dialogue history, a set of token-offset quotation spans, and an intent utterance. Building on this abstraction, we introduce a quotation-centric data pipeline that automatically synthesises task-specific dialogues, verifies answer correctness through multi-stage consistency checks, and yields both a heterogeneous training corpus and the first benchmark covering five representative scenarios. To meet the benchmark's zero-overhead and parameter-efficiency requirements, we propose QuAda, a lightweight training-based method that attaches two bottleneck projections to every attention head, dynamically amplifying or suppressing attention to quoted spans at inference time while leaving the prompt unchanged and updating < 2.8% of backbone weights. Experiments across models show that QuAda is suitable for all scenarios and generalises to unseen topics, offering an effective, plug-and-play solution for quotation-aware dialogue.
Silencer: From Discovery to Mitigation of Self-Bias in LLM-as-Benchmark-Generator
May 27, 2025



Abstract:LLM-as-Benchmark-Generator methods have been widely studied as a supplement to human annotators for scalable evaluation, while the potential biases within this paradigm remain underexplored. In this work, we systematically define and validate the phenomenon of inflated performance in models evaluated on their self-generated benchmarks, referred to as self-bias, and attribute it to sub-biases arising from question domain, language style, and wrong labels. On this basis, we propose Silencer, a general framework that leverages the heterogeneity between multiple generators at both the sample and benchmark levels to neutralize bias and generate high-quality, self-bias-silenced benchmark. Experimental results across various settings demonstrate that Silencer can suppress self-bias to near zero, significantly improve evaluation effectiveness of the generated benchmark (with an average improvement from 0.655 to 0.833 in Pearson correlation with high-quality human-annotated benchmark), while also exhibiting strong generalizability.
AdCare-VLM: Leveraging Large Vision Language Model (LVLM) to Monitor Long-Term Medication Adherence and Care
May 01, 2025Abstract:Chronic diseases, including diabetes, hypertension, asthma, HIV-AIDS, epilepsy, and tuberculosis, necessitate rigorous adherence to medication to avert disease progression, manage symptoms, and decrease mortality rates. Adherence is frequently undermined by factors including patient behavior, caregiver support, elevated medical costs, and insufficient healthcare infrastructure. We propose AdCare-VLM, a specialized Video-LLaVA-based multimodal large vision language model (LVLM) aimed at visual question answering (VQA) concerning medication adherence through patient videos. We employ a private dataset comprising 806 custom-annotated tuberculosis (TB) medication monitoring videos, which have been labeled by clinical experts, to fine-tune the model for adherence pattern detection. We present LLM-TB-VQA, a detailed medical adherence VQA dataset that encompasses positive, negative, and ambiguous adherence cases. Our method identifies correlations between visual features, such as the clear visibility of the patient's face, medication, water intake, and the act of ingestion, and their associated medical concepts in captions. This facilitates the integration of aligned visual-linguistic representations and improves multimodal interactions. Experimental results indicate that our method surpasses parameter-efficient fine-tuning (PEFT) enabled VLM models, such as LLaVA-V1.5 and Chat-UniVi, with absolute improvements ranging from 3.1% to 3.54% across pre-trained, regular, and low-rank adaptation (LoRA) configurations. Comprehensive ablation studies and attention map visualizations substantiate our approach, enhancing interpretability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge