Bin Pu
A Foundation Model for Chest X-ray Interpretation with Grounded Reasoning via Online Reinforcement Learning
Sep 04, 2025Abstract:Medical foundation models (FMs) have shown tremendous promise amid the rapid advancements in artificial intelligence (AI) technologies. However, current medical FMs typically generate answers in a black-box manner, lacking transparent reasoning processes and locally grounded interpretability, which hinders their practical clinical deployments. To this end, we introduce DeepMedix-R1, a holistic medical FM for chest X-ray (CXR) interpretation. It leverages a sequential training pipeline: initially fine-tuned on curated CXR instruction data to equip with fundamental CXR interpretation capabilities, then exposed to high-quality synthetic reasoning samples to enable cold-start reasoning, and finally refined via online reinforcement learning to enhance both grounded reasoning quality and generation performance. Thus, the model produces both an answer and reasoning steps tied to the image's local regions for each query. Quantitative evaluation demonstrates substantial improvements in report generation (e.g., 14.54% and 31.32% over LLaVA-Rad and MedGemma) and visual question answering (e.g., 57.75% and 23.06% over MedGemma and CheXagent) tasks. To facilitate robust assessment, we propose Report Arena, a benchmarking framework using advanced language models to evaluate answer quality, further highlighting the superiority of DeepMedix-R1. Expert review of generated reasoning steps reveals greater interpretability and clinical plausibility compared to the established Qwen2.5-VL-7B model (0.7416 vs. 0.2584 overall preference). Collectively, our work advances medical FM development toward holistic, transparent, and clinically actionable modeling for CXR interpretation.
Simple is what you need for efficient and accurate medical image segmentation
Jun 16, 2025Abstract:While modern segmentation models often prioritize performance over practicality, we advocate a design philosophy prioritizing simplicity and efficiency, and attempted high performance segmentation model design. This paper presents SimpleUNet, a scalable ultra-lightweight medical image segmentation model with three key innovations: (1) A partial feature selection mechanism in skip connections for redundancy reduction while enhancing segmentation performance; (2) A fixed-width architecture that prevents exponential parameter growth across network stages; (3) An adaptive feature fusion module achieving enhanced representation with minimal computational overhead. With a record-breaking 16 KB parameter configuration, SimpleUNet outperforms LBUNet and other lightweight benchmarks across multiple public datasets. The 0.67 MB variant achieves superior efficiency (8.60 GFLOPs) and accuracy, attaining a mean DSC/IoU of 85.76%/75.60% on multi-center breast lesion datasets, surpassing both U-Net and TransUNet. Evaluations on skin lesion datasets (ISIC 2017/2018: mDice 84.86%/88.77%) and endoscopic polyp segmentation (KVASIR-SEG: 86.46%/76.48% mDice/mIoU) confirm consistent dominance over state-of-the-art models. This work demonstrates that extreme model compression need not compromise performance, providing new insights for efficient and accurate medical image segmentation. Codes can be found at https://github.com/Frankyu5666666/SimpleUNet.
AI-Enabled Accurate Non-Invasive Assessment of Pulmonary Hypertension Progression via Multi-Modal Echocardiography
May 12, 2025Abstract:Echocardiographers can detect pulmonary hypertension using Doppler echocardiography; however, accurately assessing its progression often proves challenging. Right heart catheterization (RHC), the gold standard for precise evaluation, is invasive and unsuitable for routine use, limiting its practicality for timely diagnosis and monitoring of pulmonary hypertension progression. Here, we propose MePH, a multi-view, multi-modal vision-language model to accurately assess pulmonary hypertension progression using non-invasive echocardiography. We constructed a large dataset comprising paired standardized echocardiogram videos, spectral images and RHC data, covering 1,237 patient cases from 12 medical centers. For the first time, MePH precisely models the correlation between non-invasive multi-view, multi-modal echocardiography and the pressure and resistance obtained via RHC. We show that MePH significantly outperforms echocardiographers' assessments using echocardiography, reducing the mean absolute error in estimating mean pulmonary arterial pressure (mPAP) and pulmonary vascular resistance (PVR) by 49.73% and 43.81%, respectively. In eight independent external hospitals, MePH achieved a mean absolute error of 3.147 for PVR assessment. Furthermore, MePH achieved an area under the curve of 0.921, surpassing echocardiographers (area under the curve of 0.842) in accurately predicting the severity of pulmonary hypertension, whether mild or severe. A prospective study demonstrated that MePH can predict treatment efficacy for patients. Our work provides pulmonary hypertension patients with a non-invasive and timely method for monitoring disease progression, improving the accuracy and efficiency of pulmonary hypertension management while enabling earlier interventions and more personalized treatment decisions.
Test-Time Domain Generalization via Universe Learning: A Multi-Graph Matching Approach for Medical Image Segmentation
Mar 17, 2025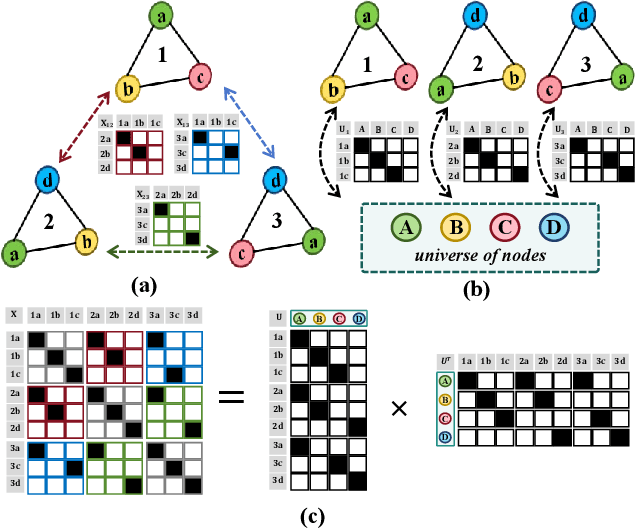
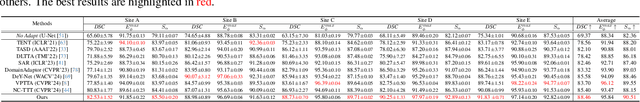
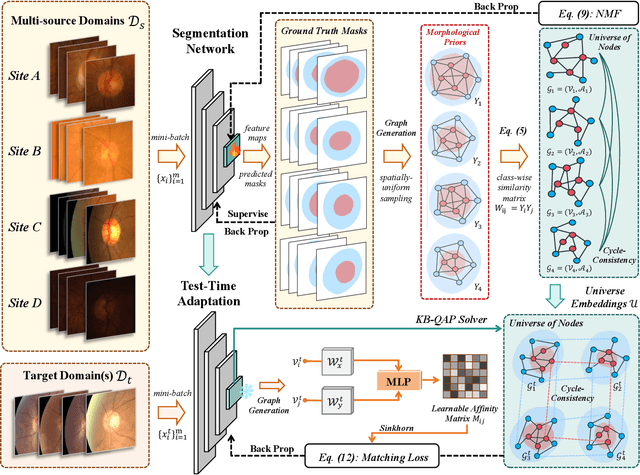
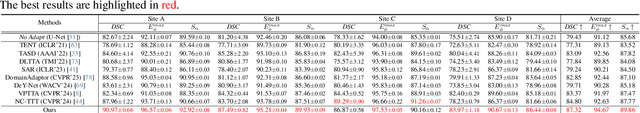
Abstract:Despite domain generalization (DG) has significantly addressed the performance degradation of pre-trained models caused by domain shifts, it often falls short in real-world deployment. Test-time adaptation (TTA), which adjusts a learned model using unlabeled test data, presents a promising solution. However, most existing TTA methods struggle to deliver strong performance in medical image segmentation, primarily because they overlook the crucial prior knowledge inherent to medical images. To address this challenge, we incorporate morphological information and propose a framework based on multi-graph matching. Specifically, we introduce learnable universe embeddings that integrate morphological priors during multi-source training, along with novel unsupervised test-time paradigms for domain adaptation. This approach guarantees cycle-consistency in multi-matching while enabling the model to more effectively capture the invariant priors of unseen data, significantly mitigating the effects of domain shifts. Extensive experiments demonstrate that our method outperforms other state-of-the-art approaches on two medical image segmentation benchmarks for both multi-source and single-source domain generalization tasks. The source code is available at https://github.com/Yore0/TTDG-MGM.
Task-Specific Knowledge Distillation from the Vision Foundation Model for Enhanced Medical Image Segmentation
Mar 10, 2025



Abstract:Large-scale pre-trained models, such as Vision Foundation Models (VFMs), have demonstrated impressive performance across various downstream tasks by transferring generalized knowledge, especially when target data is limited. However, their high computational cost and the domain gap between natural and medical images limit their practical application in medical segmentation tasks. Motivated by this, we pose the following important question: "How can we effectively utilize the knowledge of large pre-trained VFMs to train a small, task-specific model for medical image segmentation when training data is limited?" To address this problem, we propose a novel and generalizable task-specific knowledge distillation framework. Our method fine-tunes the VFM on the target segmentation task to capture task-specific features before distilling the knowledge to smaller models, leveraging Low-Rank Adaptation (LoRA) to reduce the computational cost of fine-tuning. Additionally, we incorporate synthetic data generated by diffusion models to augment the transfer set, enhancing model performance in data-limited scenarios. Experimental results across five medical image datasets demonstrate that our method consistently outperforms task-agnostic knowledge distillation and self-supervised pretraining approaches like MoCo v3 and Masked Autoencoders (MAE). For example, on the KidneyUS dataset, our method achieved a 28% higher Dice score than task-agnostic KD using 80 labeled samples for fine-tuning. On the CHAOS dataset, it achieved an 11% improvement over MAE with 100 labeled samples. These results underscore the potential of task-specific knowledge distillation to train accurate, efficient models for medical image segmentation in data-constrained settings.
Semantic Prior Distillation with Vision Foundation Model for Enhanced Rapid Bone Scintigraphy Image Restoration
Mar 04, 2025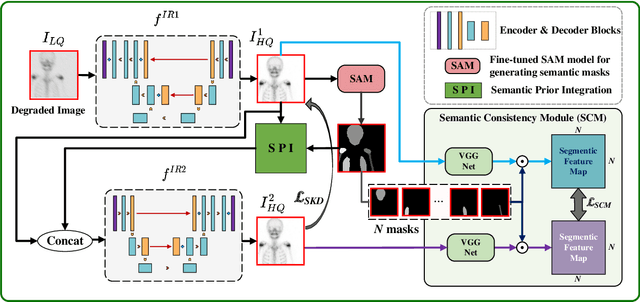
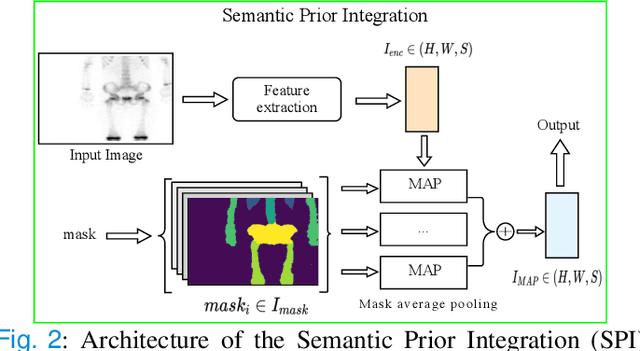
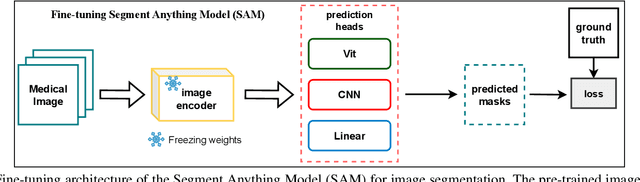
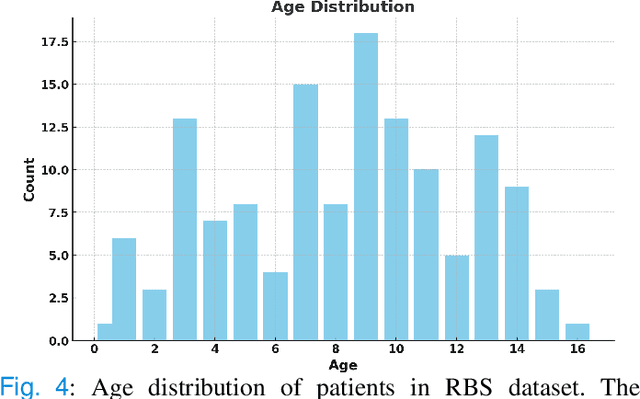
Abstract:Rapid bone scintigraphy is an essential tool for diagnosing skeletal diseases and tumor metastasis in pediatric patients, as it reduces scan time and minimizes patient discomfort. However, rapid scans often result in poor image quality, potentially affecting diagnosis due to reduced resolution and detail, which make it challenging to identify and evaluate finer anatomical structures. To address this issue, we propose the first application of SAM-based semantic priors for medical image restoration, leveraging the Segment Anything Model (SAM) to enhance rapid bone scintigraphy images in pediatric populations. Our method comprises two cascaded networks, $f^{IR1}$ and $f^{IR2}$, augmented by three key modules: a Semantic Prior Integration (SPI) module, a Semantic Knowledge Distillation (SKD) module, and a Semantic Consistency Module (SCM). The SPI and SKD modules incorporate domain-specific semantic information from a fine-tuned SAM, while the SCM maintains consistent semantic feature representation throughout the cascaded networks. In addition, we will release a novel Rapid Bone Scintigraphy dataset called RBS, the first dataset dedicated to rapid bone scintigraphy image restoration in pediatric patients. RBS consists of 137 pediatric patients aged between 0.5 and 16 years who underwent both standard and rapid bone scans. The dataset includes scans performed at 20 cm/min (standard) and 40 cm/min (rapid), representing a $2\times$ acceleration. We conducted extensive experiments on both the publicly available endoscopic dataset and RBS. The results demonstrate that our method outperforms all existing methods across various metrics, including PSNR, SSIM, FID, and LPIPS.
Vision Foundation Models in Medical Image Analysis: Advances and Challenges
Feb 21, 2025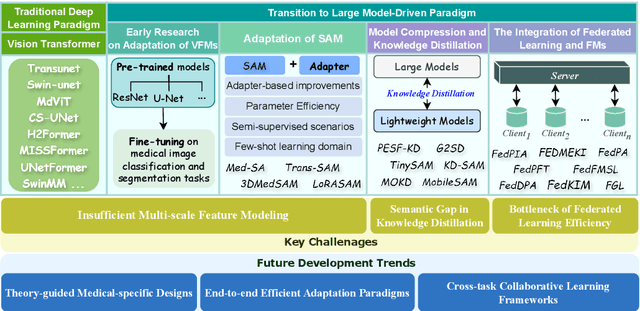
Abstract:The rapid development of Vision Foundation Models (VFMs), particularly Vision Transformers (ViT) and Segment Anything Model (SAM), has sparked significant advances in the field of medical image analysis. These models have demonstrated exceptional capabilities in capturing long-range dependencies and achieving high generalization in segmentation tasks. However, adapting these large models to medical image analysis presents several challenges, including domain differences between medical and natural images, the need for efficient model adaptation strategies, and the limitations of small-scale medical datasets. This paper reviews the state-of-the-art research on the adaptation of VFMs to medical image segmentation, focusing on the challenges of domain adaptation, model compression, and federated learning. We discuss the latest developments in adapter-based improvements, knowledge distillation techniques, and multi-scale contextual feature modeling, and propose future directions to overcome these bottlenecks. Our analysis highlights the potential of VFMs, along with emerging methodologies such as federated learning and model compression, to revolutionize medical image analysis and enhance clinical applications. The goal of this work is to provide a comprehensive overview of current approaches and suggest key areas for future research that can drive the next wave of innovation in medical image segmentation.
Topology-Aware Wavelet Mamba for Airway Structure Segmentation in Postoperative Recurrent Nasopharyngeal Carcinoma CT Scans
Feb 20, 2025Abstract:Nasopharyngeal carcinoma (NPC) patients often undergo radiotherapy and chemotherapy, which can lead to postoperative complications such as limited mouth opening and joint stiffness, particularly in recurrent cases that require re-surgery. These complications can affect airway function, making accurate postoperative airway risk assessment essential for managing patient care. Accurate segmentation of airway-related structures in postoperative CT scans is crucial for assessing these risks. This study introduces TopoWMamba (Topology-aware Wavelet Mamba), a novel segmentation model specifically designed to address the challenges of postoperative airway risk evaluation in recurrent NPC patients. TopoWMamba combines wavelet-based multi-scale feature extraction, state-space sequence modeling, and topology-aware modules to segment airway-related structures in CT scans robustly. By leveraging the Wavelet-based Mamba Block (WMB) for hierarchical frequency decomposition and the Snake Conv VSS (SCVSS) module to preserve anatomical continuity, TopoWMamba effectively captures both fine-grained boundaries and global structural context, crucial for accurate segmentation in complex postoperative scenarios. Through extensive testing on the NPCSegCT dataset, TopoWMamba achieves an average Dice score of 88.02%, outperforming existing models such as UNet, Attention UNet, and SwinUNet. Additionally, TopoWMamba is tested on the SegRap 2023 Challenge dataset, where it shows a significant improvement in trachea segmentation with a Dice score of 95.26%. The proposed model provides a strong foundation for automated segmentation, enabling more accurate postoperative airway risk evaluation.
CardiacNet: Learning to Reconstruct Abnormalities for Cardiac Disease Assessment from Echocardiogram Videos
Oct 28, 2024Abstract:Echocardiogram video plays a crucial role in analysing cardiac function and diagnosing cardiac diseases. Current deep neural network methods primarily aim to enhance diagnosis accuracy by incorporating prior knowledge, such as segmenting cardiac structures or lesions annotated by human experts. However, diagnosing the inconsistent behaviours of the heart, which exist across both spatial and temporal dimensions, remains extremely challenging. For instance, the analysis of cardiac motion acquires both spatial and temporal information from the heartbeat cycle. To address this issue, we propose a novel reconstruction-based approach named CardiacNet to learn a better representation of local cardiac structures and motion abnormalities through echocardiogram videos. CardiacNet is accompanied by the Consistency Deformation Codebook (CDC) and the Consistency Deformed-Discriminator (CDD) to learn the commonalities across abnormal and normal samples by incorporating cardiac prior knowledge. In addition, we propose benchmark datasets named CardiacNet-PAH and CardiacNet-ASD to evaluate the effectiveness of cardiac disease assessment. In experiments, our CardiacNet can achieve state-of-the-art results in three different cardiac disease assessment tasks on public datasets CAMUS, EchoNet, and our datasets. The code and dataset are available at: https://github.com/xmed-lab/CardiacNet.
Bidirectional Recurrence for Cardiac Motion Tracking with Gaussian Process Latent Coding
Oct 28, 2024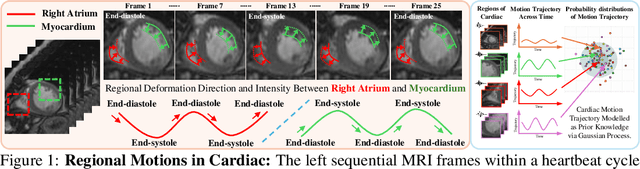
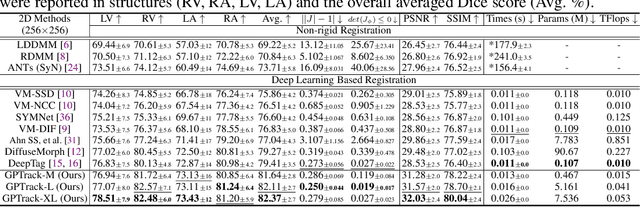
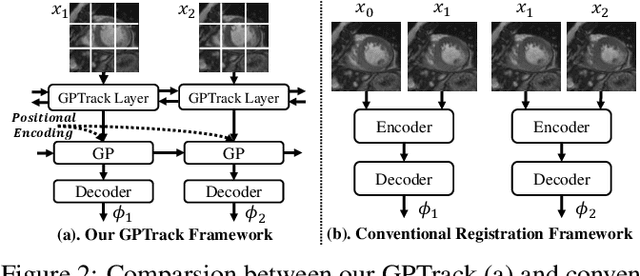
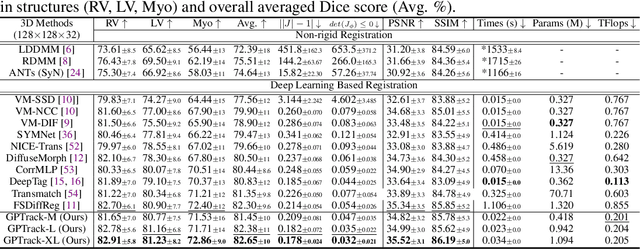
Abstract:Quantitative analysis of cardiac motion is crucial for assessing cardiac function. This analysis typically uses imaging modalities such as MRI and Echocardiograms that capture detailed image sequences throughout the heartbeat cycle. Previous methods predominantly focused on the analysis of image pairs lacking consideration of the motion dynamics and spatial variability. Consequently, these methods often overlook the long-term relationships and regional motion characteristic of cardiac. To overcome these limitations, we introduce the \textbf{GPTrack}, a novel unsupervised framework crafted to fully explore the temporal and spatial dynamics of cardiac motion. The GPTrack enhances motion tracking by employing the sequential Gaussian Process in the latent space and encoding statistics by spatial information at each time stamp, which robustly promotes temporal consistency and spatial variability of cardiac dynamics. Also, we innovatively aggregate sequential information in a bidirectional recursive manner, mimicking the behavior of diffeomorphic registration to better capture consistent long-term relationships of motions across cardiac regions such as the ventricles and atria. Our GPTrack significantly improves the precision of motion tracking in both 3D and 4D medical images while maintaining computational efficiency. The code is available at: https://github.com/xmed-lab/GPTrack
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge