Pengchen Liang
MS-UMamba: An Improved Vision Mamba Unet for Fetal Abdominal Medical Image Segmentation
Jun 14, 2025Abstract:Recently, Mamba-based methods have become popular in medical image segmentation due to their lightweight design and long-range dependency modeling capabilities. However, current segmentation methods frequently encounter challenges in fetal ultrasound images, such as enclosed anatomical structures, blurred boundaries, and small anatomical structures. To address the need for balancing local feature extraction and global context modeling, we propose MS-UMamba, a novel hybrid convolutional-mamba model for fetal ultrasound image segmentation. Specifically, we design a visual state space block integrated with a CNN branch (SS-MCAT-SSM), which leverages Mamba's global modeling strengths and convolutional layers' local representation advantages to enhance feature learning. In addition, we also propose an efficient multi-scale feature fusion module that integrates spatial attention mechanisms, which Integrating feature information from different layers enhances the feature representation ability of the model. Finally, we conduct extensive experiments on a non-public dataset, experimental results demonstrate that MS-UMamba model has excellent performance in segmentation performance.
DCD: A Semantic Segmentation Model for Fetal Ultrasound Four-Chamber View
Jun 10, 2025Abstract:Accurate segmentation of anatomical structures in the apical four-chamber (A4C) view of fetal echocardiography is essential for early diagnosis and prenatal evaluation of congenital heart disease (CHD). However, precise segmentation remains challenging due to ultrasound artifacts, speckle noise, anatomical variability, and boundary ambiguity across different gestational stages. To reduce the workload of sonographers and enhance segmentation accuracy, we propose DCD, an advanced deep learning-based model for automatic segmentation of key anatomical structures in the fetal A4C view. Our model incorporates a Dense Atrous Spatial Pyramid Pooling (Dense ASPP) module, enabling superior multi-scale feature extraction, and a Convolutional Block Attention Module (CBAM) to enhance adaptive feature representation. By effectively capturing both local and global contextual information, DCD achieves precise and robust segmentation, contributing to improved prenatal cardiac assessment.
Task-Specific Knowledge Distillation from the Vision Foundation Model for Enhanced Medical Image Segmentation
Mar 10, 2025



Abstract:Large-scale pre-trained models, such as Vision Foundation Models (VFMs), have demonstrated impressive performance across various downstream tasks by transferring generalized knowledge, especially when target data is limited. However, their high computational cost and the domain gap between natural and medical images limit their practical application in medical segmentation tasks. Motivated by this, we pose the following important question: "How can we effectively utilize the knowledge of large pre-trained VFMs to train a small, task-specific model for medical image segmentation when training data is limited?" To address this problem, we propose a novel and generalizable task-specific knowledge distillation framework. Our method fine-tunes the VFM on the target segmentation task to capture task-specific features before distilling the knowledge to smaller models, leveraging Low-Rank Adaptation (LoRA) to reduce the computational cost of fine-tuning. Additionally, we incorporate synthetic data generated by diffusion models to augment the transfer set, enhancing model performance in data-limited scenarios. Experimental results across five medical image datasets demonstrate that our method consistently outperforms task-agnostic knowledge distillation and self-supervised pretraining approaches like MoCo v3 and Masked Autoencoders (MAE). For example, on the KidneyUS dataset, our method achieved a 28% higher Dice score than task-agnostic KD using 80 labeled samples for fine-tuning. On the CHAOS dataset, it achieved an 11% improvement over MAE with 100 labeled samples. These results underscore the potential of task-specific knowledge distillation to train accurate, efficient models for medical image segmentation in data-constrained settings.
Semantic Prior Distillation with Vision Foundation Model for Enhanced Rapid Bone Scintigraphy Image Restoration
Mar 04, 2025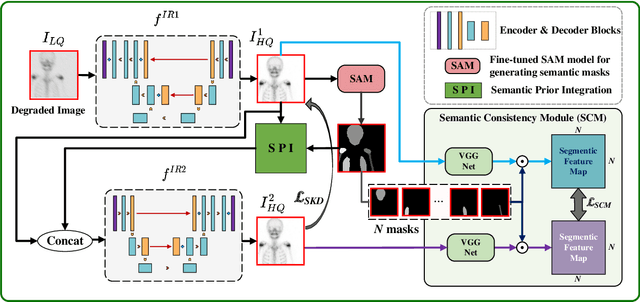
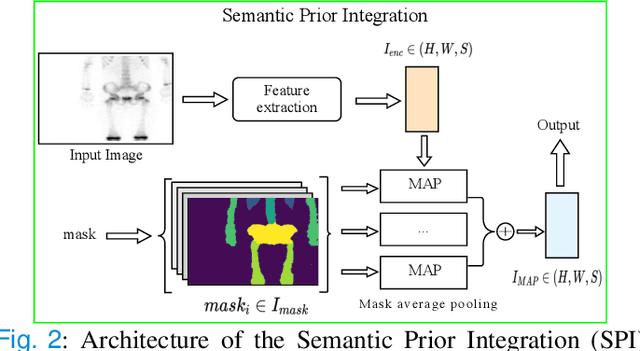
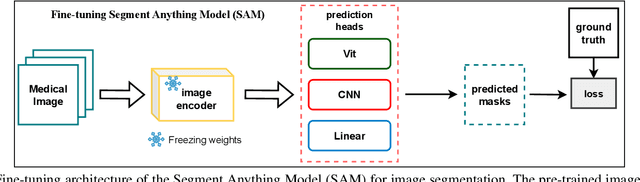
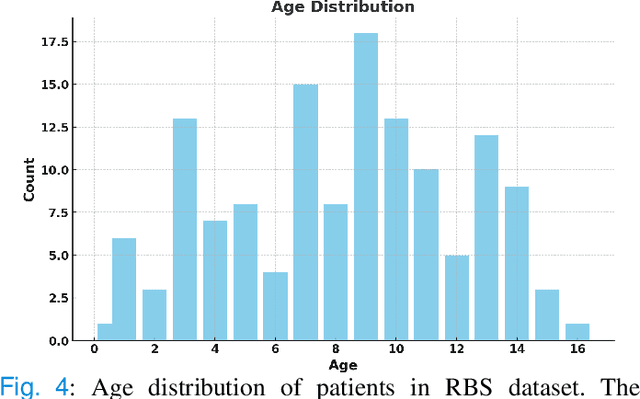
Abstract:Rapid bone scintigraphy is an essential tool for diagnosing skeletal diseases and tumor metastasis in pediatric patients, as it reduces scan time and minimizes patient discomfort. However, rapid scans often result in poor image quality, potentially affecting diagnosis due to reduced resolution and detail, which make it challenging to identify and evaluate finer anatomical structures. To address this issue, we propose the first application of SAM-based semantic priors for medical image restoration, leveraging the Segment Anything Model (SAM) to enhance rapid bone scintigraphy images in pediatric populations. Our method comprises two cascaded networks, $f^{IR1}$ and $f^{IR2}$, augmented by three key modules: a Semantic Prior Integration (SPI) module, a Semantic Knowledge Distillation (SKD) module, and a Semantic Consistency Module (SCM). The SPI and SKD modules incorporate domain-specific semantic information from a fine-tuned SAM, while the SCM maintains consistent semantic feature representation throughout the cascaded networks. In addition, we will release a novel Rapid Bone Scintigraphy dataset called RBS, the first dataset dedicated to rapid bone scintigraphy image restoration in pediatric patients. RBS consists of 137 pediatric patients aged between 0.5 and 16 years who underwent both standard and rapid bone scans. The dataset includes scans performed at 20 cm/min (standard) and 40 cm/min (rapid), representing a $2\times$ acceleration. We conducted extensive experiments on both the publicly available endoscopic dataset and RBS. The results demonstrate that our method outperforms all existing methods across various metrics, including PSNR, SSIM, FID, and LPIPS.
Vision Foundation Models in Medical Image Analysis: Advances and Challenges
Feb 21, 2025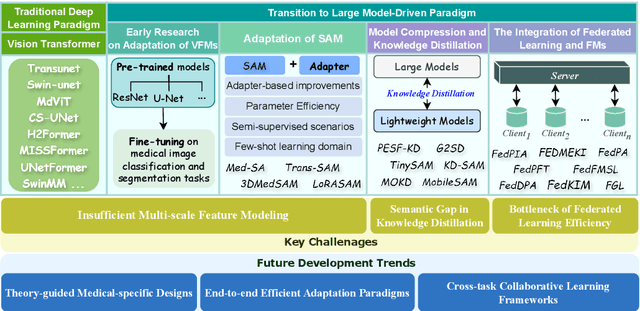
Abstract:The rapid development of Vision Foundation Models (VFMs), particularly Vision Transformers (ViT) and Segment Anything Model (SAM), has sparked significant advances in the field of medical image analysis. These models have demonstrated exceptional capabilities in capturing long-range dependencies and achieving high generalization in segmentation tasks. However, adapting these large models to medical image analysis presents several challenges, including domain differences between medical and natural images, the need for efficient model adaptation strategies, and the limitations of small-scale medical datasets. This paper reviews the state-of-the-art research on the adaptation of VFMs to medical image segmentation, focusing on the challenges of domain adaptation, model compression, and federated learning. We discuss the latest developments in adapter-based improvements, knowledge distillation techniques, and multi-scale contextual feature modeling, and propose future directions to overcome these bottlenecks. Our analysis highlights the potential of VFMs, along with emerging methodologies such as federated learning and model compression, to revolutionize medical image analysis and enhance clinical applications. The goal of this work is to provide a comprehensive overview of current approaches and suggest key areas for future research that can drive the next wave of innovation in medical image segmentation.
Topology-Aware Wavelet Mamba for Airway Structure Segmentation in Postoperative Recurrent Nasopharyngeal Carcinoma CT Scans
Feb 20, 2025Abstract:Nasopharyngeal carcinoma (NPC) patients often undergo radiotherapy and chemotherapy, which can lead to postoperative complications such as limited mouth opening and joint stiffness, particularly in recurrent cases that require re-surgery. These complications can affect airway function, making accurate postoperative airway risk assessment essential for managing patient care. Accurate segmentation of airway-related structures in postoperative CT scans is crucial for assessing these risks. This study introduces TopoWMamba (Topology-aware Wavelet Mamba), a novel segmentation model specifically designed to address the challenges of postoperative airway risk evaluation in recurrent NPC patients. TopoWMamba combines wavelet-based multi-scale feature extraction, state-space sequence modeling, and topology-aware modules to segment airway-related structures in CT scans robustly. By leveraging the Wavelet-based Mamba Block (WMB) for hierarchical frequency decomposition and the Snake Conv VSS (SCVSS) module to preserve anatomical continuity, TopoWMamba effectively captures both fine-grained boundaries and global structural context, crucial for accurate segmentation in complex postoperative scenarios. Through extensive testing on the NPCSegCT dataset, TopoWMamba achieves an average Dice score of 88.02%, outperforming existing models such as UNet, Attention UNet, and SwinUNet. Additionally, TopoWMamba is tested on the SegRap 2023 Challenge dataset, where it shows a significant improvement in trachea segmentation with a Dice score of 95.26%. The proposed model provides a strong foundation for automated segmentation, enabling more accurate postoperative airway risk evaluation.
Toward Zero-Shot Learning for Visual Dehazing of Urological Surgical Robots
Oct 02, 2024



Abstract:Robot-assisted surgery has profoundly influenced current forms of minimally invasive surgery. However, in transurethral suburethral urological surgical robots, they need to work in a liquid environment. This causes vaporization of the liquid when shearing and heating is performed, resulting in bubble atomization that affects the visual perception of the robot. This can lead to the need for uninterrupted pauses in the surgical procedure, which makes the surgery take longer. To address the atomization characteristics of liquids under urological surgical robotic vision, we propose an unsupervised zero-shot dehaze method (RSF-Dehaze) for urological surgical robotic vision. Specifically, the proposed Region Similarity Filling Module (RSFM) of RSF-Dehaze significantly improves the recovery of blurred region tissues. In addition, we organize and propose a dehaze dataset for robotic vision in urological surgery (USRobot-Dehaze dataset). In particular, this dataset contains the three most common urological surgical robot operation scenarios. To the best of our knowledge, we are the first to organize and propose a publicly available dehaze dataset for urological surgical robot vision. The proposed RSF-Dehaze proves the effectiveness of our method in three urological surgical robot operation scenarios with extensive comparative experiments with 20 most classical and advanced dehazing and image recovery algorithms. The proposed source code and dataset are available at https://github.com/wurenkai/RSF-Dehaze .
UltraLight VM-UNet: Parallel Vision Mamba Significantly Reduces Parameters for Skin Lesion Segmentation
Apr 09, 2024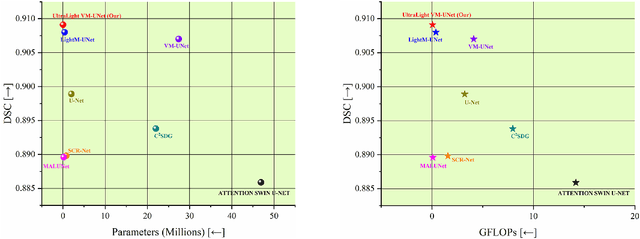

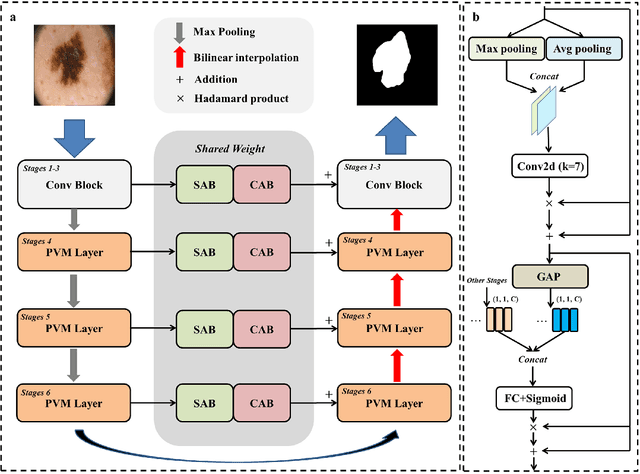
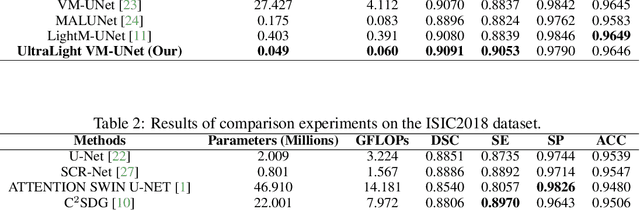
Abstract:Traditionally for improving the segmentation performance of models, most approaches prefer to use adding more complex modules. And this is not suitable for the medical field, especially for mobile medical devices, where computationally loaded models are not suitable for real clinical environments due to computational resource constraints. Recently, state-space models (SSMs), represented by Mamba, have become a strong competitor to traditional CNNs and Transformers. In this paper, we deeply explore the key elements of parameter influence in Mamba and propose an UltraLight Vision Mamba UNet (UltraLight VM-UNet) based on this. Specifically, we propose a method for processing features in parallel Vision Mamba, named PVM Layer, which achieves excellent performance with the lowest computational load while keeping the overall number of processing channels constant. We conducted comparisons and ablation experiments with several state-of-the-art lightweight models on three skin lesion public datasets and demonstrated that the UltraLight VM-UNet exhibits the same strong performance competitiveness with parameters of only 0.049M and GFLOPs of 0.060. In addition, this study deeply explores the key elements of parameter influence in Mamba, which will lay a theoretical foundation for Mamba to possibly become a new mainstream module for lightweighting in the future. The code is available from https://github.com/wurenkai/UltraLight-VM-UNet .
H-vmunet: High-order Vision Mamba UNet for Medical Image Segmentation
Mar 20, 2024



Abstract:In the field of medical image segmentation, variant models based on Convolutional Neural Networks (CNNs) and Visual Transformers (ViTs) as the base modules have been very widely developed and applied. However, CNNs are often limited in their ability to deal with long sequences of information, while the low sensitivity of ViTs to local feature information and the problem of secondary computational complexity limit their development. Recently, the emergence of state-space models (SSMs), especially 2D-selective-scan (SS2D), has had an impact on the longtime dominance of traditional CNNs and ViTs as the foundational modules of visual neural networks. In this paper, we extend the adaptability of SS2D by proposing a High-order Vision Mamba UNet (H-vmunet) for medical image segmentation. Among them, the proposed High-order 2D-selective-scan (H-SS2D) progressively reduces the introduction of redundant information during SS2D operations through higher-order interactions. In addition, the proposed Local-SS2D module improves the learning ability of local features of SS2D at each order of interaction. We conducted comparison and ablation experiments on three publicly available medical image datasets (ISIC2017, Spleen, and CVC-ClinicDB), and the results all demonstrate the strong competitiveness of H-vmunet in medical image segmentation tasks. The code is available from https://github.com/wurenkai/H-vmunet .
Only Positive Cases: 5-fold High-order Attention Interaction Model for Skin Segmentation Derived Classification
Nov 27, 2023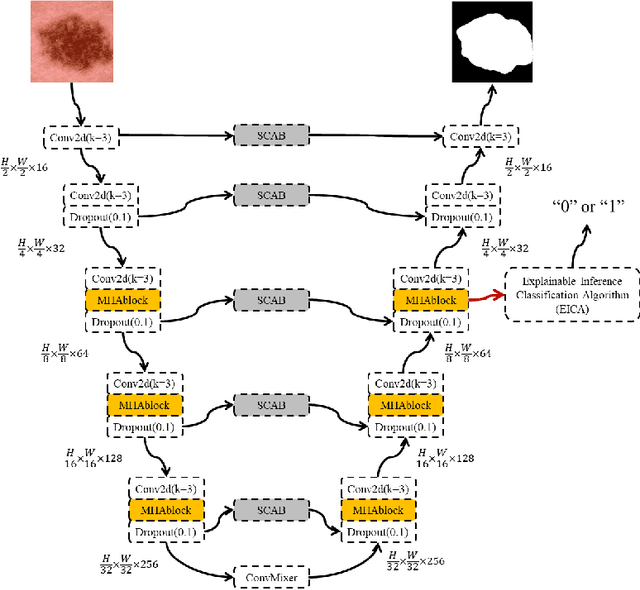
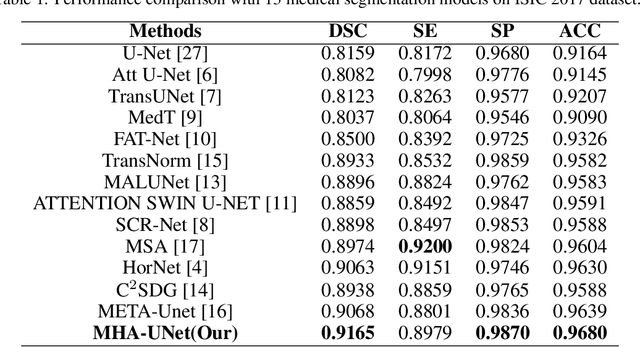
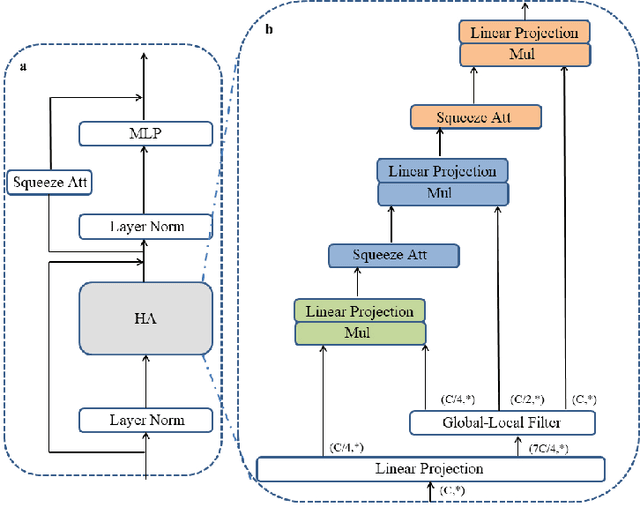

Abstract:Computer-aided diagnosis of skin diseases is an important tool. However, the interpretability of computer-aided diagnosis is currently poor. Dermatologists and patients cannot intuitively understand the learning and prediction process of neural networks, which will lead to a decrease in the credibility of computer-aided diagnosis. In addition, traditional methods need to be trained using negative samples in order to predict the presence or absence of a lesion, but medical data is often in short supply. In this paper, we propose a multiple high-order attention interaction model (MHA-UNet) for use in a highly explainable skin lesion segmentation task. MHA-UNet is able to obtain the presence or absence of a lesion by explainable reasoning without the need for training on negative samples. Specifically, we propose a high-order attention interaction mechanism that introduces squeeze attention to a higher level for feature attention. In addition, a multiple high-order attention interaction (MHAblock) module is proposed by combining the different features of different orders. For classifying the presence or absence of lesions, we conducted classification experiments on several publicly available datasets in the absence of negative samples, based on explainable reasoning about the interaction of 5 attention orders of MHAblock. The highest positive detection rate obtained from the experiments was 81.0% and the highest negative detection rate was 83.5%. For segmentation experiments, comparison experiments of the proposed method with 13 medical segmentation models and external validation experiments with 8 state-of-the-art models in three public datasets and our clinical dataset demonstrate the state-of-the-art performance of our model. The code is available from https://github.com/wurenkai/MHA-UNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge