Peilong Wang
Fine-Tuning Open-Source Large Language Models to Improve Their Performance on Radiation Oncology Tasks: A Feasibility Study to Investigate Their Potential Clinical Applications in Radiation Oncology
Jan 28, 2025



Abstract:Background: The radiation oncology clinical practice involves many steps relying on the dynamic interplay of abundant text data. Large language models have displayed remarkable capabilities in processing complex text information. But their direct applications in specific fields like radiation oncology remain underexplored. Purpose: This study aims to investigate whether fine-tuning LLMs with domain knowledge can improve the performance on Task (1) treatment regimen generation, Task (2) treatment modality selection (photon, proton, electron, or brachytherapy), and Task (3) ICD-10 code prediction in radiation oncology. Methods: Data for 15,724 patient cases were extracted. Cases where patients had a single diagnostic record, and a clearly identifiable primary treatment plan were selected for preprocessing and manual annotation to have 7,903 cases of the patient diagnosis, treatment plan, treatment modality, and ICD-10 code. Each case was used to construct a pair consisting of patient diagnostics details and an answer (treatment regimen, treatment modality, or ICD-10 code respectively) for the supervised fine-tuning of these three tasks. Open source LLaMA2-7B and Mistral-7B models were utilized for the fine-tuning with the Low-Rank Approximations method. Accuracy and ROUGE-1 score were reported for the fine-tuned models and original models. Clinical evaluation was performed on Task (1) by radiation oncologists, while precision, recall, and F-1 score were evaluated for Task (2) and (3). One-sided Wilcoxon signed-rank tests were used to statistically analyze the results. Results: Fine-tuned LLMs outperformed original LLMs across all tasks with p-value <= 0.001. Clinical evaluation demonstrated that over 60% of the fine-tuned LLMs-generated treatment regimens were clinically acceptable. Precision, recall, and F1-score showed improved performance of fine-tuned LLMs.
Evaluating The Performance of Using Large Language Models to Automate Summarization of CT Simulation Orders in Radiation Oncology
Jan 27, 2025



Abstract:Purpose: This study aims to use a large language model (LLM) to automate the generation of summaries from the CT simulation orders and evaluate its performance. Materials and Methods: A total of 607 CT simulation orders for patients were collected from the Aria database at our institution. A locally hosted Llama 3.1 405B model, accessed via the Application Programming Interface (API) service, was used to extract keywords from the CT simulation orders and generate summaries. The downloaded CT simulation orders were categorized into seven groups based on treatment modalities and disease sites. For each group, a customized instruction prompt was developed collaboratively with therapists to guide the Llama 3.1 405B model in generating summaries. The ground truth for the corresponding summaries was manually derived by carefully reviewing each CT simulation order and subsequently verified by therapists. The accuracy of the LLM-generated summaries was evaluated by therapists using the verified ground truth as a reference. Results: About 98% of the LLM-generated summaries aligned with the manually generated ground truth in terms of accuracy. Our evaluations showed an improved consistency in format and enhanced readability of the LLM-generated summaries compared to the corresponding therapists-generated summaries. This automated approach demonstrated a consistent performance across all groups, regardless of modality or disease site. Conclusions: This study demonstrated the high precision and consistency of the Llama 3.1 405B model in extracting keywords and summarizing CT simulation orders, suggesting that LLMs have great potential to help with this task, reduce the workload of therapists and improve workflow efficiency.
Large Language Models for Bioinformatics
Jan 10, 2025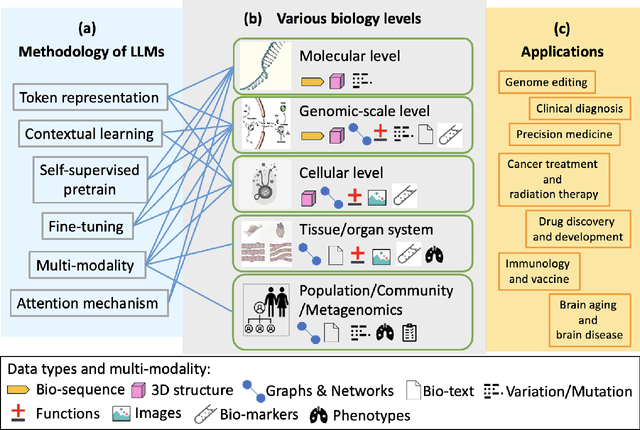
Abstract:With the rapid advancements in large language model (LLM) technology and the emergence of bioinformatics-specific language models (BioLMs), there is a growing need for a comprehensive analysis of the current landscape, computational characteristics, and diverse applications. This survey aims to address this need by providing a thorough review of BioLMs, focusing on their evolution, classification, and distinguishing features, alongside a detailed examination of training methodologies, datasets, and evaluation frameworks. We explore the wide-ranging applications of BioLMs in critical areas such as disease diagnosis, drug discovery, and vaccine development, highlighting their impact and transformative potential in bioinformatics. We identify key challenges and limitations inherent in BioLMs, including data privacy and security concerns, interpretability issues, biases in training data and model outputs, and domain adaptation complexities. Finally, we highlight emerging trends and future directions, offering valuable insights to guide researchers and clinicians toward advancing BioLMs for increasingly sophisticated biological and clinical applications.
A recent evaluation on the performance of LLMs on radiation oncology physics using questions of randomly shuffled options
Dec 14, 2024



Abstract:Purpose: We present an updated study evaluating the performance of large language models (LLMs) in answering radiation oncology physics questions, focusing on the latest released models. Methods: A set of 100 multiple-choice radiation oncology physics questions, previously created by us, was used for this study. The answer options of the questions were randomly shuffled to create "new" exam sets. Five LLMs -- OpenAI o1-preview, GPT-4o, LLaMA 3.1 (405B), Gemini 1.5 Pro, and Claude 3.5 Sonnet -- with the versions released before September 30, 2024, were queried using these new exams. To evaluate their deductive reasoning abilities, the correct answer options in the questions were replaced with "None of the above." Then, the explain-first and step-by-step instruction prompt was used to test if it improved their reasoning abilities. The performance of the LLMs was compared to medical physicists in majority-vote scenarios. Results: All models demonstrated expert-level performance on these questions, with o1-preview even surpassing medical physicists in majority-vote scenarios. When substituting the correct answer options with "None of the above," all models exhibited a considerable decline in performance, suggesting room for improvement. The explain-first and step-by-step instruction prompt helped enhance the reasoning abilities of LLaMA 3.1 (405B), Gemini 1.5 Pro, and Claude 3.5 Sonnet models. Conclusion: These latest LLMs demonstrated expert-level performance in answering radiation oncology physics questions, exhibiting great potential for assisting in radiation oncology physics education.
Evaluation of OpenAI o1: Opportunities and Challenges of AGI
Sep 27, 2024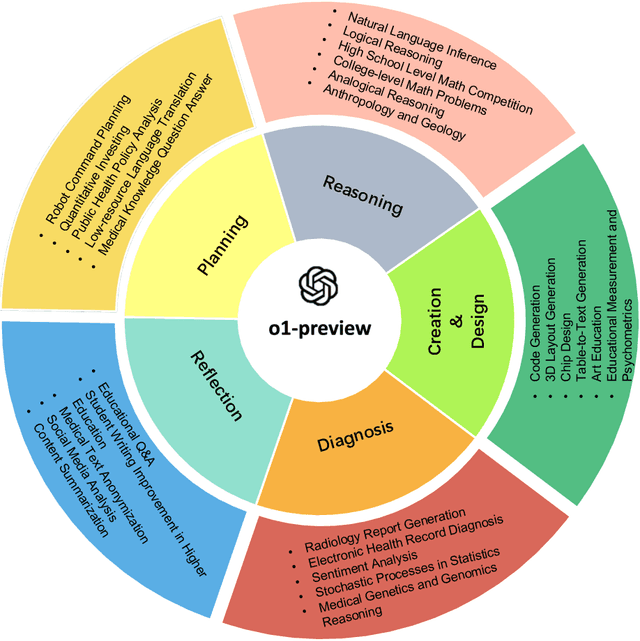
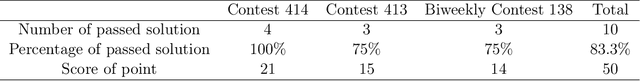
Abstract:This comprehensive study evaluates the performance of OpenAI's o1-preview large language model across a diverse array of complex reasoning tasks, spanning multiple domains, including computer science, mathematics, natural sciences, medicine, linguistics, and social sciences. Through rigorous testing, o1-preview demonstrated remarkable capabilities, often achieving human-level or superior performance in areas ranging from coding challenges to scientific reasoning and from language processing to creative problem-solving. Key findings include: -83.3% success rate in solving complex competitive programming problems, surpassing many human experts. -Superior ability in generating coherent and accurate radiology reports, outperforming other evaluated models. -100% accuracy in high school-level mathematical reasoning tasks, providing detailed step-by-step solutions. -Advanced natural language inference capabilities across general and specialized domains like medicine. -Impressive performance in chip design tasks, outperforming specialized models in areas such as EDA script generation and bug analysis. -Remarkable proficiency in anthropology and geology, demonstrating deep understanding and reasoning in these specialized fields. -Strong capabilities in quantitative investing. O1 has comprehensive financial knowledge and statistical modeling skills. -Effective performance in social media analysis, including sentiment analysis and emotion recognition. The model excelled particularly in tasks requiring intricate reasoning and knowledge integration across various fields. While some limitations were observed, including occasional errors on simpler problems and challenges with certain highly specialized concepts, the overall results indicate significant progress towards artificial general intelligence.
A Classification-Based Adaptive Segmentation Pipeline: Feasibility Study Using Polycystic Liver Disease and Metastases from Colorectal Cancer CT Images
May 02, 2024Abstract:Automated segmentation tools often encounter accuracy and adaptability issues when applied to images of different pathology. The purpose of this study is to explore the feasibility of building a workflow to efficiently route images to specifically trained segmentation models. By implementing a deep learning classifier to automatically classify the images and route them to appropriate segmentation models, we hope that our workflow can segment the images with different pathology accurately. The data we used in this study are 350 CT images from patients affected by polycystic liver disease and 350 CT images from patients presenting with liver metastases from colorectal cancer. All images had the liver manually segmented by trained imaging analysts. Our proposed adaptive segmentation workflow achieved a statistically significant improvement for the task of total liver segmentation compared to the generic single segmentation model (non-parametric Wilcoxon signed rank test, n=100, p-value << 0.001). This approach is applicable in a wide range of scenarios and should prove useful in clinical implementations of segmentation pipelines.
The Radiation Oncology NLP Database
Jan 19, 2024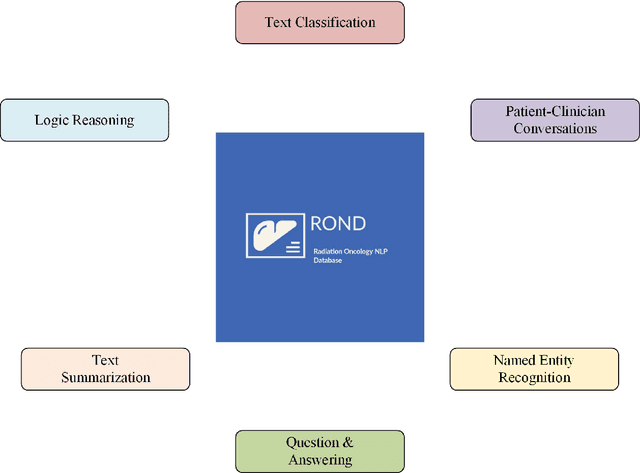

Abstract:We present the Radiation Oncology NLP Database (ROND), the first dedicated Natural Language Processing (NLP) dataset for radiation oncology, an important medical specialty that has received limited attention from the NLP community in the past. With the advent of Artificial General Intelligence (AGI), there is an increasing need for specialized datasets and benchmarks to facilitate research and development. ROND is specifically designed to address this gap in the domain of radiation oncology, a field that offers many opportunities for NLP exploration. It encompasses various NLP tasks including Logic Reasoning, Text Classification, Named Entity Recognition (NER), Question Answering (QA), Text Summarization, and Patient-Clinician Conversations, each with a distinct focus on radiation oncology concepts and application cases. In addition, we have developed an instruction-tuning dataset consisting of over 20k instruction pairs (based on ROND) and trained a large language model, CancerChat. This serves to demonstrate the potential of instruction-tuning large language models within a highly-specialized medical domain. The evaluation results in this study could serve as baseline results for future research. ROND aims to stimulate advancements in radiation oncology and clinical NLP by offering a platform for testing and improving algorithms and models in a domain-specific context. The ROND dataset is a joint effort of multiple U.S. health institutions. The data is available at https://github.com/zl-liu/Radiation-Oncology-NLP-Database.
RadOnc-GPT: A Large Language Model for Radiation Oncology
Sep 22, 2023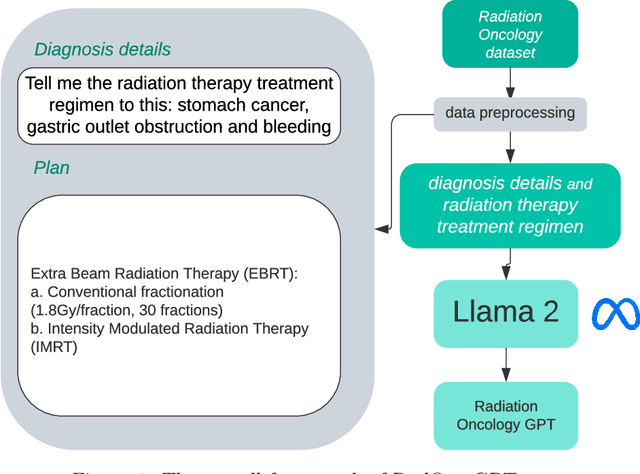
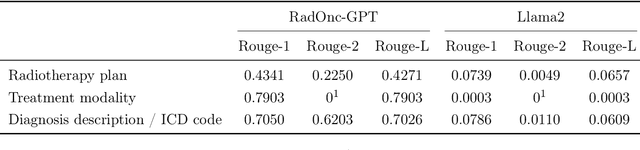
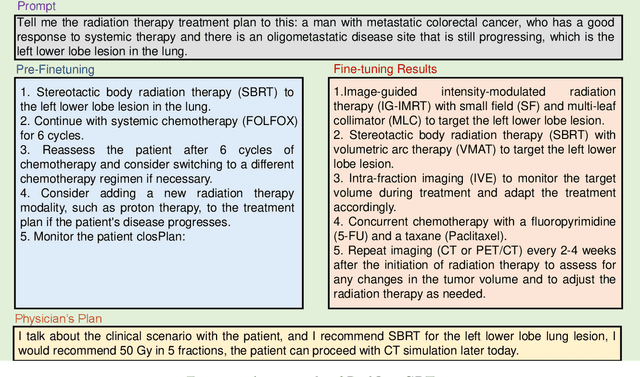
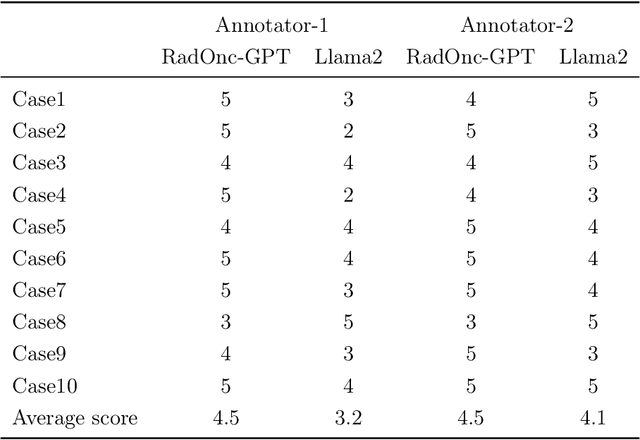
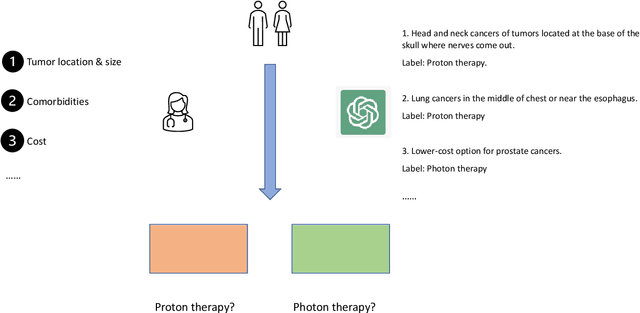
Abstract:This paper presents RadOnc-GPT, a large language model specialized for radiation oncology through advanced tuning methods. RadOnc-GPT was finetuned on a large dataset of radiation oncology patient records and clinical notes from the Mayo Clinic in Arizona. The model employs instruction tuning on three key tasks - generating radiotherapy treatment regimens, determining optimal radiation modalities, and providing diagnostic descriptions/ICD codes based on patient diagnostic details. Evaluations conducted by comparing RadOnc-GPT outputs to general large language model outputs showed that RadOnc-GPT generated outputs with significantly improved clarity, specificity, and clinical relevance. The study demonstrated the potential of using large language models fine-tuned using domain-specific knowledge like RadOnc-GPT to achieve transformational capabilities in highly specialized healthcare fields such as radiation oncology.
Evaluating Large Language Models for Radiology Natural Language Processing
Jul 27, 2023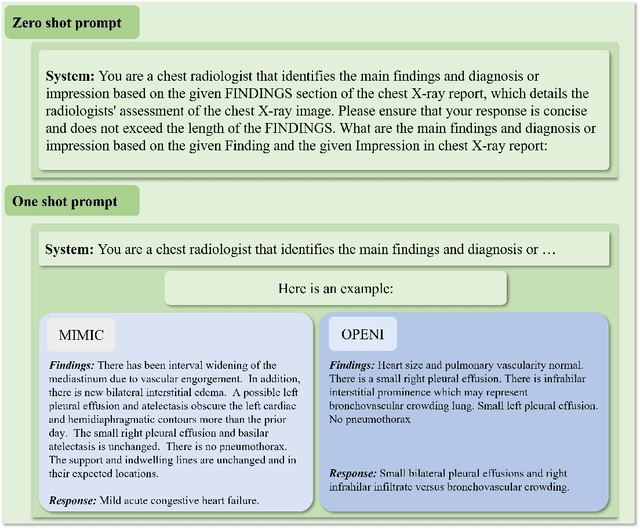
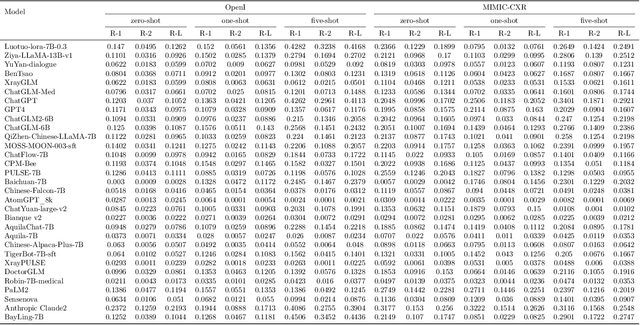
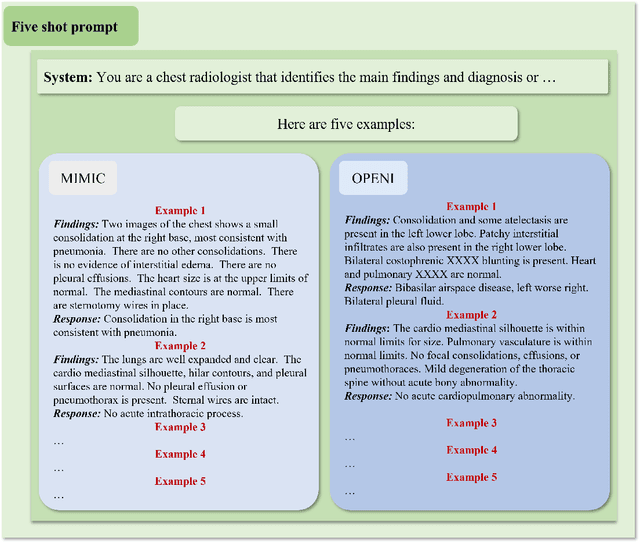
Abstract:The rise of large language models (LLMs) has marked a pivotal shift in the field of natural language processing (NLP). LLMs have revolutionized a multitude of domains, and they have made a significant impact in the medical field. Large language models are now more abundant than ever, and many of these models exhibit bilingual capabilities, proficient in both English and Chinese. However, a comprehensive evaluation of these models remains to be conducted. This lack of assessment is especially apparent within the context of radiology NLP. This study seeks to bridge this gap by critically evaluating thirty two LLMs in interpreting radiology reports, a crucial component of radiology NLP. Specifically, the ability to derive impressions from radiologic findings is assessed. The outcomes of this evaluation provide key insights into the performance, strengths, and weaknesses of these LLMs, informing their practical applications within the medical domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge