Chenbin Liu
TG-LMM: Enhancing Medical Image Segmentation Accuracy through Text-Guided Large Multi-Modal Model
Sep 05, 2024



Abstract:We propose TG-LMM (Text-Guided Large Multi-Modal Model), a novel approach that leverages textual descriptions of organs to enhance segmentation accuracy in medical images. Existing medical image segmentation methods face several challenges: current medical automatic segmentation models do not effectively utilize prior knowledge, such as descriptions of organ locations; previous text-visual models focus on identifying the target rather than improving the segmentation accuracy; prior models attempt to use prior knowledge to enhance accuracy but do not incorporate pre-trained models. To address these issues, TG-LMM integrates prior knowledge, specifically expert descriptions of the spatial locations of organs, into the segmentation process. Our model utilizes pre-trained image and text encoders to reduce the number of training parameters and accelerate the training process. Additionally, we designed a comprehensive image-text information fusion structure to ensure thorough integration of the two modalities of data. We evaluated TG-LMM on three authoritative medical image datasets, encompassing the segmentation of various parts of the human body. Our method demonstrated superior performance compared to existing approaches, such as MedSAM, SAM and nnUnet.
Quality assurance of organs-at-risk delineation in radiotherapy
May 20, 2024



Abstract:The delineation of tumor target and organs-at-risk is critical in the radiotherapy treatment planning. Automatic segmentation can be used to reduce the physician workload and improve the consistency. However, the quality assurance of the automatic segmentation is still an unmet need in clinical practice. The patient data used in our study was a standardized dataset from AAPM Thoracic Auto-Segmentation Challenge. The OARs included were left and right lungs, heart, esophagus, and spinal cord. Two groups of OARs were generated, the benchmark dataset manually contoured by experienced physicians and the test dataset automatically created using a software AccuContour. A resnet-152 network was performed as feature extractor, and one-class support vector classifier was used to determine the high or low quality. We evaluate the model performance with balanced accuracy, F-score, sensitivity, specificity and the area under the receiving operator characteristic curve. We randomly generated contour errors to assess the generalization of our method, explored the detection limit, and evaluated the correlations between detection limit and various metrics such as volume, Dice similarity coefficient, Hausdorff distance, and mean surface distance. The proposed one-class classifier outperformed in metrics such as balanced accuracy, AUC, and others. The proposed method showed significant improvement over binary classifiers in handling various types of errors. Our proposed model, which introduces residual network and attention mechanism in the one-class classification framework, was able to detect the various types of OAR contour errors with high accuracy. The proposed method can significantly reduce the burden of physician review for contour delineation.
The Radiation Oncology NLP Database
Jan 19, 2024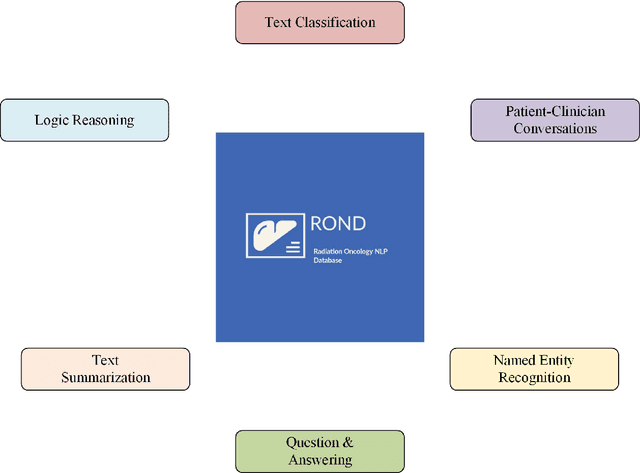
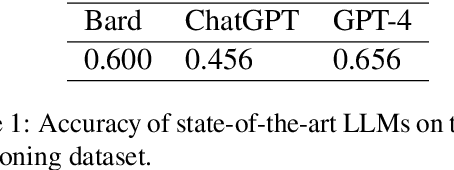

Abstract:We present the Radiation Oncology NLP Database (ROND), the first dedicated Natural Language Processing (NLP) dataset for radiation oncology, an important medical specialty that has received limited attention from the NLP community in the past. With the advent of Artificial General Intelligence (AGI), there is an increasing need for specialized datasets and benchmarks to facilitate research and development. ROND is specifically designed to address this gap in the domain of radiation oncology, a field that offers many opportunities for NLP exploration. It encompasses various NLP tasks including Logic Reasoning, Text Classification, Named Entity Recognition (NER), Question Answering (QA), Text Summarization, and Patient-Clinician Conversations, each with a distinct focus on radiation oncology concepts and application cases. In addition, we have developed an instruction-tuning dataset consisting of over 20k instruction pairs (based on ROND) and trained a large language model, CancerChat. This serves to demonstrate the potential of instruction-tuning large language models within a highly-specialized medical domain. The evaluation results in this study could serve as baseline results for future research. ROND aims to stimulate advancements in radiation oncology and clinical NLP by offering a platform for testing and improving algorithms and models in a domain-specific context. The ROND dataset is a joint effort of multiple U.S. health institutions. The data is available at https://github.com/zl-liu/Radiation-Oncology-NLP-Database.
RadOnc-GPT: A Large Language Model for Radiation Oncology
Sep 22, 2023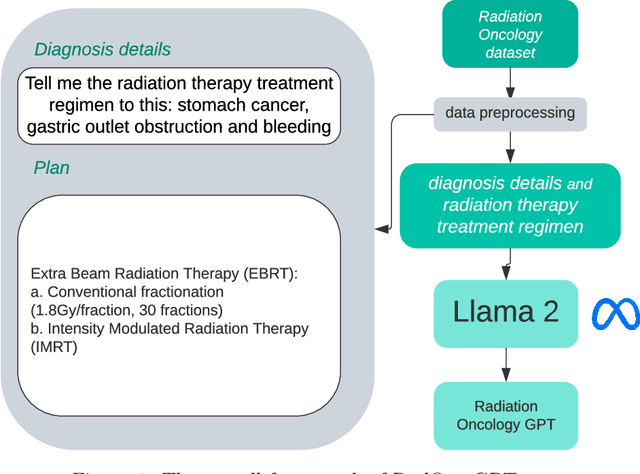
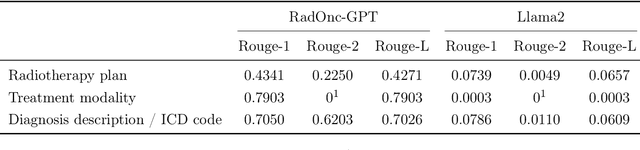
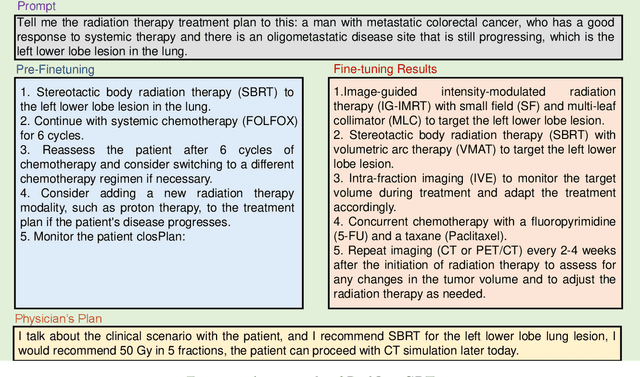
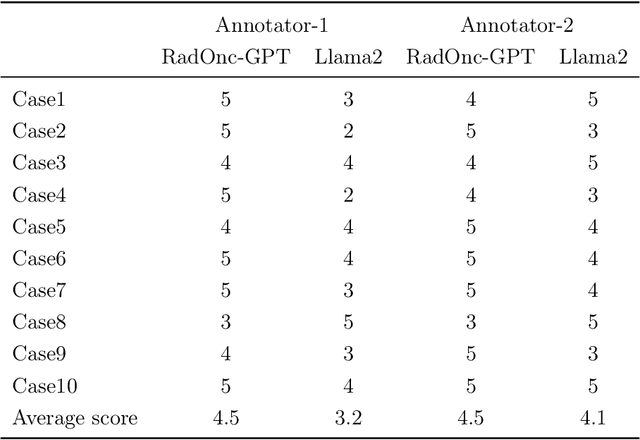
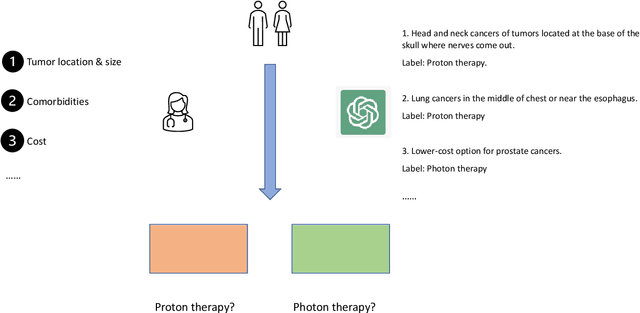
Abstract:This paper presents RadOnc-GPT, a large language model specialized for radiation oncology through advanced tuning methods. RadOnc-GPT was finetuned on a large dataset of radiation oncology patient records and clinical notes from the Mayo Clinic in Arizona. The model employs instruction tuning on three key tasks - generating radiotherapy treatment regimens, determining optimal radiation modalities, and providing diagnostic descriptions/ICD codes based on patient diagnostic details. Evaluations conducted by comparing RadOnc-GPT outputs to general large language model outputs showed that RadOnc-GPT generated outputs with significantly improved clarity, specificity, and clinical relevance. The study demonstrated the potential of using large language models fine-tuned using domain-specific knowledge like RadOnc-GPT to achieve transformational capabilities in highly specialized healthcare fields such as radiation oncology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge