Dajiang Zhu
Community-Level Modeling of Gyral Folding Patterns for Robust and Anatomically Informed Individualized Brain Mapping
Feb 01, 2026Abstract:Cortical folding exhibits substantial inter-individual variability while preserving stable anatomical landmarks that enable fine-scale characterization of cortical organization. Among these, the three-hinge gyrus (3HG) serves as a key folding primitive, showing consistent topology yet meaningful variations in morphology, connectivity, and function. Existing landmark-based methods typically model each 3HG independently, ignoring that 3HGs form higher-order folding communities that capture mesoscale structure. This simplification weakens anatomical representation and makes one-to-one matching sensitive to positional variability and noise. We propose a spectral graph representation learning framework that models community-level folding units rather than isolated landmarks. Each 3HG is encoded using a dual-profile representation combining surface topology and structural connectivity. Subject-specific spectral clustering identifies coherent folding communities, followed by topological refinement to preserve anatomical continuity. For cross-subject correspondence, we introduce Joint Morphological-Geometric Matching, jointly optimizing geometric and morphometric similarity. Across over 1000 Human Connectome Project subjects, the resulting communities show reduced morphometric variance, stronger modular organization, improved hemispheric consistency, and superior alignment compared with atlas-based and landmark-based or embedding-based baselines. These findings demonstrate that community-level modeling provides a robust and anatomically grounded framework for individualized cortical characterization and reliable cross-subject correspondence.
DCMM-Transformer: Degree-Corrected Mixed-Membership Attention for Medical Imaging
Nov 15, 2025Abstract:Medical images exhibit latent anatomical groupings, such as organs, tissues, and pathological regions, that standard Vision Transformers (ViTs) fail to exploit. While recent work like SBM-Transformer attempts to incorporate such structures through stochastic binary masking, they suffer from non-differentiability, training instability, and the inability to model complex community structure. We present DCMM-Transformer, a novel ViT architecture for medical image analysis that incorporates a Degree-Corrected Mixed-Membership (DCMM) model as an additive bias in self-attention. Unlike prior approaches that rely on multiplicative masking and binary sampling, our method introduces community structure and degree heterogeneity in a fully differentiable and interpretable manner. Comprehensive experiments across diverse medical imaging datasets, including brain, chest, breast, and ocular modalities, demonstrate the superior performance and generalizability of the proposed approach. Furthermore, the learned group structure and structured attention modulation substantially enhance interpretability by yielding attention maps that are anatomically meaningful and semantically coherent.
Knowledge Distillation and Dataset Distillation of Large Language Models: Emerging Trends, Challenges, and Future Directions
Apr 20, 2025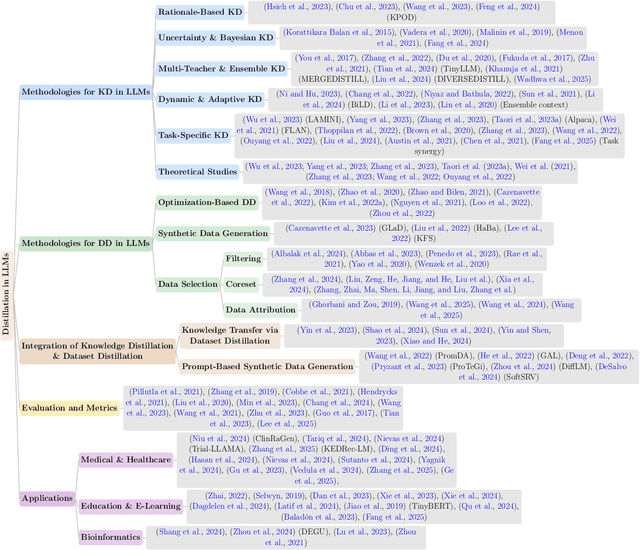
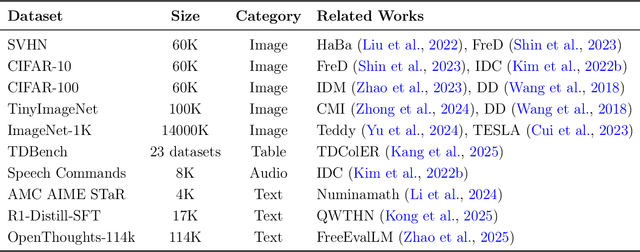
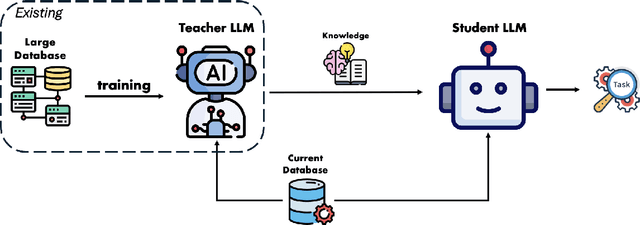
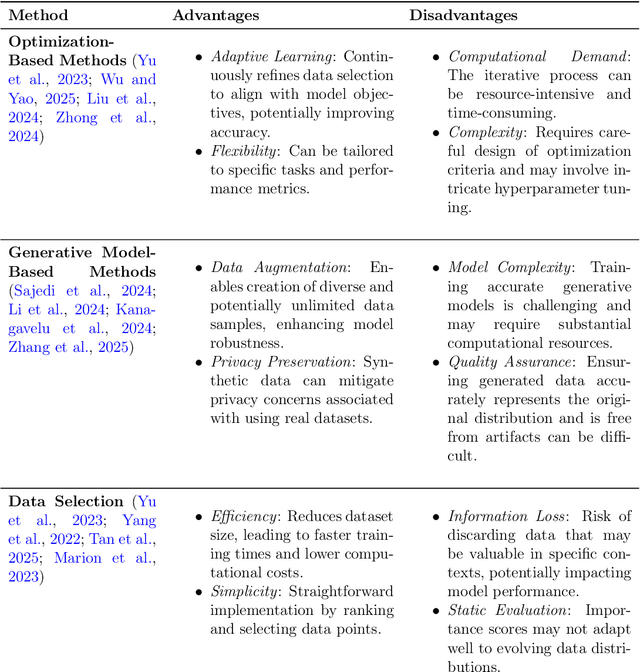
Abstract:The exponential growth of Large Language Models (LLMs) continues to highlight the need for efficient strategies to meet ever-expanding computational and data demands. This survey provides a comprehensive analysis of two complementary paradigms: Knowledge Distillation (KD) and Dataset Distillation (DD), both aimed at compressing LLMs while preserving their advanced reasoning capabilities and linguistic diversity. We first examine key methodologies in KD, such as task-specific alignment, rationale-based training, and multi-teacher frameworks, alongside DD techniques that synthesize compact, high-impact datasets through optimization-based gradient matching, latent space regularization, and generative synthesis. Building on these foundations, we explore how integrating KD and DD can produce more effective and scalable compression strategies. Together, these approaches address persistent challenges in model scalability, architectural heterogeneity, and the preservation of emergent LLM abilities. We further highlight applications across domains such as healthcare and education, where distillation enables efficient deployment without sacrificing performance. Despite substantial progress, open challenges remain in preserving emergent reasoning and linguistic diversity, enabling efficient adaptation to continually evolving teacher models and datasets, and establishing comprehensive evaluation protocols. By synthesizing methodological innovations, theoretical foundations, and practical insights, our survey charts a path toward sustainable, resource-efficient LLMs through the tighter integration of KD and DD principles.
AD-GPT: Large Language Models in Alzheimer's Disease
Apr 03, 2025Abstract:Large language models (LLMs) have emerged as powerful tools for medical information retrieval, yet their accuracy and depth remain limited in specialized domains such as Alzheimer's disease (AD), a growing global health challenge. To address this gap, we introduce AD-GPT, a domain-specific generative pre-trained transformer designed to enhance the retrieval and analysis of AD-related genetic and neurobiological information. AD-GPT integrates diverse biomedical data sources, including potential AD-associated genes, molecular genetic information, and key gene variants linked to brain regions. We develop a stacked LLM architecture combining Llama3 and BERT, optimized for four critical tasks in AD research: (1) genetic information retrieval, (2) gene-brain region relationship assessment, (3) gene-AD relationship analysis, and (4) brain region-AD relationship mapping. Comparative evaluations against state-of-the-art LLMs demonstrate AD-GPT's superior precision and reliability across these tasks, underscoring its potential as a robust and specialized AI tool for advancing AD research and biomarker discovery.
GyralNet Subnetwork Partitioning via Differentiable Spectral Modularity Optimization
Mar 25, 2025



Abstract:Understanding the structural and functional organization of the human brain requires a detailed examination of cortical folding patterns, among which the three-hinge gyrus (3HG) has been identified as a key structural landmark. GyralNet, a network representation of cortical folding, models 3HGs as nodes and gyral crests as edges, highlighting their role as critical hubs in cortico-cortical connectivity. However, existing methods for analyzing 3HGs face significant challenges, including the sub-voxel scale of 3HGs at typical neuroimaging resolutions, the computational complexity of establishing cross-subject correspondences, and the oversimplification of treating 3HGs as independent nodes without considering their community-level relationships. To address these limitations, we propose a fully differentiable subnetwork partitioning framework that employs a spectral modularity maximization optimization strategy to modularize the organization of 3HGs within GyralNet. By incorporating topological structural similarity and DTI-derived connectivity patterns as attribute features, our approach provides a biologically meaningful representation of cortical organization. Extensive experiments on the Human Connectome Project (HCP) dataset demonstrate that our method effectively partitions GyralNet at the individual level while preserving the community-level consistency of 3HGs across subjects, offering a robust foundation for understanding brain connectivity.
Core-Periphery Principle Guided State Space Model for Functional Connectome Classification
Mar 18, 2025Abstract:Understanding the organization of human brain networks has become a central focus in neuroscience, particularly in the study of functional connectivity, which plays a crucial role in diagnosing neurological disorders. Advances in functional magnetic resonance imaging and machine learning techniques have significantly improved brain network analysis. However, traditional machine learning approaches struggle to capture the complex relationships between brain regions, while deep learning methods, particularly Transformer-based models, face computational challenges due to their quadratic complexity in long-sequence modeling. To address these limitations, we propose a Core-Periphery State-Space Model (CP-SSM), an innovative framework for functional connectome classification. Specifically, we introduce Mamba, a selective state-space model with linear complexity, to effectively capture long-range dependencies in functional brain networks. Furthermore, inspired by the core-periphery (CP) organization, a fundamental characteristic of brain networks that enhances efficient information transmission, we design CP-MoE, a CP-guided Mixture-of-Experts that improves the representation learning of brain connectivity patterns. We evaluate CP-SSM on two benchmark fMRI datasets: ABIDE and ADNI. Experimental results demonstrate that CP-SSM surpasses Transformer-based models in classification performance while significantly reducing computational complexity. These findings highlight the effectiveness and efficiency of CP-SSM in modeling brain functional connectivity, offering a promising direction for neuroimaging-based neurological disease diagnosis.
Classification of Mild Cognitive Impairment Based on Dynamic Functional Connectivity Using Spatio-Temporal Transformer
Jan 27, 2025Abstract:Dynamic functional connectivity (dFC) using resting-state functional magnetic resonance imaging (rs-fMRI) is an advanced technique for capturing the dynamic changes of neural activities, and can be very useful in the studies of brain diseases such as Alzheimer's disease (AD). Yet, existing studies have not fully leveraged the sequential information embedded within dFC that can potentially provide valuable information when identifying brain conditions. In this paper, we propose a novel framework that jointly learns the embedding of both spatial and temporal information within dFC based on the transformer architecture. Specifically, we first construct dFC networks from rs-fMRI data through a sliding window strategy. Then, we simultaneously employ a temporal block and a spatial block to capture higher-order representations of dynamic spatio-temporal dependencies, via mapping them into an efficient fused feature representation. To further enhance the robustness of these feature representations by reducing the dependency on labeled data, we also introduce a contrastive learning strategy to manipulate different brain states. Experimental results on 345 subjects with 570 scans from the Alzheimer's Disease Neuroimaging Initiative (ADNI) demonstrate the superiority of our proposed method for MCI (Mild Cognitive Impairment, the prodromal stage of AD) prediction, highlighting its potential for early identification of AD.
Brain-Adapter: Enhancing Neurological Disorder Analysis with Adapter-Tuning Multimodal Large Language Models
Jan 27, 2025


Abstract:Understanding brain disorders is crucial for accurate clinical diagnosis and treatment. Recent advances in Multimodal Large Language Models (MLLMs) offer a promising approach to interpreting medical images with the support of text descriptions. However, previous research has primarily focused on 2D medical images, leaving richer spatial information of 3D images under-explored, and single-modality-based methods are limited by overlooking the critical clinical information contained in other modalities. To address this issue, this paper proposes Brain-Adapter, a novel approach that incorporates an extra bottleneck layer to learn new knowledge and instill it into the original pre-trained knowledge. The major idea is to incorporate a lightweight bottleneck layer to train fewer parameters while capturing essential information and utilize a Contrastive Language-Image Pre-training (CLIP) strategy to align multimodal data within a unified representation space. Extensive experiments demonstrated the effectiveness of our approach in integrating multimodal data to significantly improve the diagnosis accuracy without high computational costs, highlighting the potential to enhance real-world diagnostic workflows.
Large Language Models for Bioinformatics
Jan 10, 2025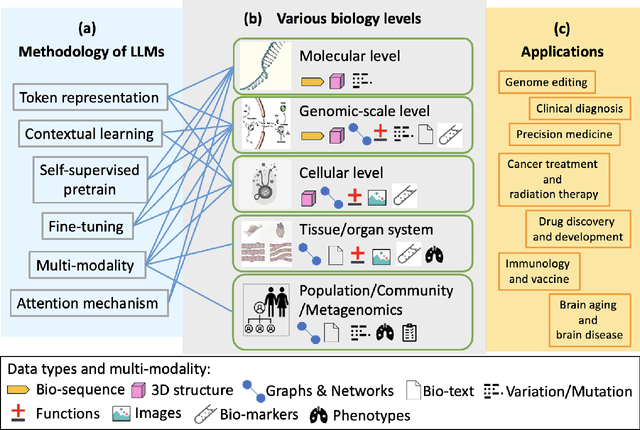
Abstract:With the rapid advancements in large language model (LLM) technology and the emergence of bioinformatics-specific language models (BioLMs), there is a growing need for a comprehensive analysis of the current landscape, computational characteristics, and diverse applications. This survey aims to address this need by providing a thorough review of BioLMs, focusing on their evolution, classification, and distinguishing features, alongside a detailed examination of training methodologies, datasets, and evaluation frameworks. We explore the wide-ranging applications of BioLMs in critical areas such as disease diagnosis, drug discovery, and vaccine development, highlighting their impact and transformative potential in bioinformatics. We identify key challenges and limitations inherent in BioLMs, including data privacy and security concerns, interpretability issues, biases in training data and model outputs, and domain adaptation complexities. Finally, we highlight emerging trends and future directions, offering valuable insights to guide researchers and clinicians toward advancing BioLMs for increasingly sophisticated biological and clinical applications.
Towards Next-Generation Medical Agent: How o1 is Reshaping Decision-Making in Medical Scenarios
Nov 16, 2024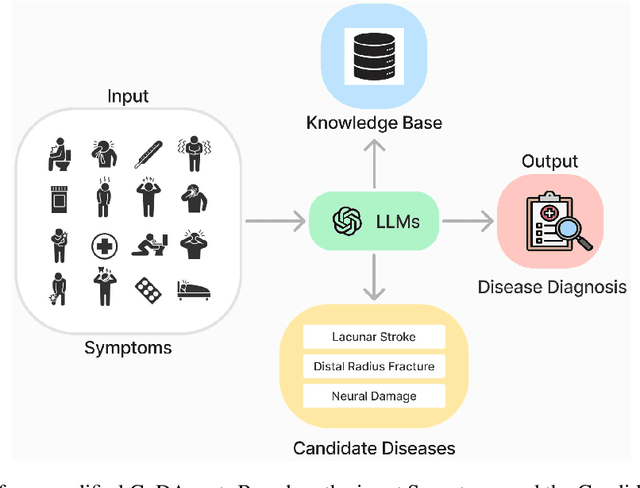
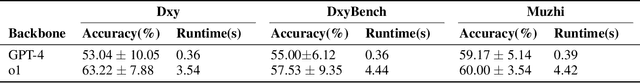
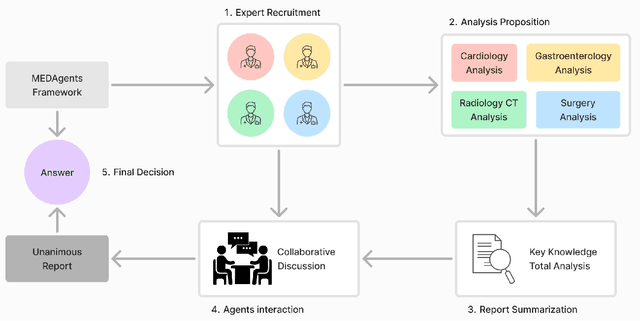

Abstract:Artificial Intelligence (AI) has become essential in modern healthcare, with large language models (LLMs) offering promising advances in clinical decision-making. Traditional model-based approaches, including those leveraging in-context demonstrations and those with specialized medical fine-tuning, have demonstrated strong performance in medical language processing but struggle with real-time adaptability, multi-step reasoning, and handling complex medical tasks. Agent-based AI systems address these limitations by incorporating reasoning traces, tool selection based on context, knowledge retrieval, and both short- and long-term memory. These additional features enable the medical AI agent to handle complex medical scenarios where decision-making should be built on real-time interaction with the environment. Therefore, unlike conventional model-based approaches that treat medical queries as isolated questions, medical AI agents approach them as complex tasks and behave more like human doctors. In this paper, we study the choice of the backbone LLM for medical AI agents, which is the foundation for the agent's overall reasoning and action generation. In particular, we consider the emergent o1 model and examine its impact on agents' reasoning, tool-use adaptability, and real-time information retrieval across diverse clinical scenarios, including high-stakes settings such as intensive care units (ICUs). Our findings demonstrate o1's ability to enhance diagnostic accuracy and consistency, paving the way for smarter, more responsive AI tools that support better patient outcomes and decision-making efficacy in clinical practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge