Chao Cao
Community-Level Modeling of Gyral Folding Patterns for Robust and Anatomically Informed Individualized Brain Mapping
Feb 01, 2026Abstract:Cortical folding exhibits substantial inter-individual variability while preserving stable anatomical landmarks that enable fine-scale characterization of cortical organization. Among these, the three-hinge gyrus (3HG) serves as a key folding primitive, showing consistent topology yet meaningful variations in morphology, connectivity, and function. Existing landmark-based methods typically model each 3HG independently, ignoring that 3HGs form higher-order folding communities that capture mesoscale structure. This simplification weakens anatomical representation and makes one-to-one matching sensitive to positional variability and noise. We propose a spectral graph representation learning framework that models community-level folding units rather than isolated landmarks. Each 3HG is encoded using a dual-profile representation combining surface topology and structural connectivity. Subject-specific spectral clustering identifies coherent folding communities, followed by topological refinement to preserve anatomical continuity. For cross-subject correspondence, we introduce Joint Morphological-Geometric Matching, jointly optimizing geometric and morphometric similarity. Across over 1000 Human Connectome Project subjects, the resulting communities show reduced morphometric variance, stronger modular organization, improved hemispheric consistency, and superior alignment compared with atlas-based and landmark-based or embedding-based baselines. These findings demonstrate that community-level modeling provides a robust and anatomically grounded framework for individualized cortical characterization and reliable cross-subject correspondence.
RPT*: Global Planning with Probabilistic Terminals for Target Search in Complex Environments
Jan 19, 2026Abstract:Routing problems such as Hamiltonian Path Problem (HPP), seeks a path to visit all the vertices in a graph while minimizing the path cost. This paper studies a variant, HPP with Probabilistic Terminals (HPP-PT), where each vertex has a probability representing the likelihood that the robot's path terminates there, and the objective is to minimize the expected path cost. HPP-PT arises in target object search, where a mobile robot must visit all candidate locations to find an object, and prior knowledge of the object's location is expressed as vertex probabilities. While routing problems have been studied for decades, few of them consider uncertainty as required in this work. The challenge lies not only in optimally ordering the vertices, as in standard HPP, but also in handling history dependency: the expected path cost depends on the order in which vertices were previously visited. This makes many existing methods inefficient or inapplicable. To address the challenge, we propose a search-based approach RPT* with solution optimality guarantees, which leverages dynamic programming in a new state space to bypass the history dependency and novel heuristics to speed up the computation. Building on RPT*, we design a Hierarchical Autonomous Target Search (HATS) system that combines RPT* with either Bayesian filtering for lifelong target search with noisy sensors, or autonomous exploration to find targets in unknown environments. Experiments in both simulation and real robot show that our approach can naturally balance between exploitation and exploration, thereby finding targets more quickly on average than baseline methods.
DCMM-Transformer: Degree-Corrected Mixed-Membership Attention for Medical Imaging
Nov 15, 2025Abstract:Medical images exhibit latent anatomical groupings, such as organs, tissues, and pathological regions, that standard Vision Transformers (ViTs) fail to exploit. While recent work like SBM-Transformer attempts to incorporate such structures through stochastic binary masking, they suffer from non-differentiability, training instability, and the inability to model complex community structure. We present DCMM-Transformer, a novel ViT architecture for medical image analysis that incorporates a Degree-Corrected Mixed-Membership (DCMM) model as an additive bias in self-attention. Unlike prior approaches that rely on multiplicative masking and binary sampling, our method introduces community structure and degree heterogeneity in a fully differentiable and interpretable manner. Comprehensive experiments across diverse medical imaging datasets, including brain, chest, breast, and ocular modalities, demonstrate the superior performance and generalizability of the proposed approach. Furthermore, the learned group structure and structured attention modulation substantially enhance interpretability by yielding attention maps that are anatomically meaningful and semantically coherent.
GyralNet Subnetwork Partitioning via Differentiable Spectral Modularity Optimization
Mar 25, 2025



Abstract:Understanding the structural and functional organization of the human brain requires a detailed examination of cortical folding patterns, among which the three-hinge gyrus (3HG) has been identified as a key structural landmark. GyralNet, a network representation of cortical folding, models 3HGs as nodes and gyral crests as edges, highlighting their role as critical hubs in cortico-cortical connectivity. However, existing methods for analyzing 3HGs face significant challenges, including the sub-voxel scale of 3HGs at typical neuroimaging resolutions, the computational complexity of establishing cross-subject correspondences, and the oversimplification of treating 3HGs as independent nodes without considering their community-level relationships. To address these limitations, we propose a fully differentiable subnetwork partitioning framework that employs a spectral modularity maximization optimization strategy to modularize the organization of 3HGs within GyralNet. By incorporating topological structural similarity and DTI-derived connectivity patterns as attribute features, our approach provides a biologically meaningful representation of cortical organization. Extensive experiments on the Human Connectome Project (HCP) dataset demonstrate that our method effectively partitions GyralNet at the individual level while preserving the community-level consistency of 3HGs across subjects, offering a robust foundation for understanding brain connectivity.
Core-Periphery Principle Guided State Space Model for Functional Connectome Classification
Mar 18, 2025Abstract:Understanding the organization of human brain networks has become a central focus in neuroscience, particularly in the study of functional connectivity, which plays a crucial role in diagnosing neurological disorders. Advances in functional magnetic resonance imaging and machine learning techniques have significantly improved brain network analysis. However, traditional machine learning approaches struggle to capture the complex relationships between brain regions, while deep learning methods, particularly Transformer-based models, face computational challenges due to their quadratic complexity in long-sequence modeling. To address these limitations, we propose a Core-Periphery State-Space Model (CP-SSM), an innovative framework for functional connectome classification. Specifically, we introduce Mamba, a selective state-space model with linear complexity, to effectively capture long-range dependencies in functional brain networks. Furthermore, inspired by the core-periphery (CP) organization, a fundamental characteristic of brain networks that enhances efficient information transmission, we design CP-MoE, a CP-guided Mixture-of-Experts that improves the representation learning of brain connectivity patterns. We evaluate CP-SSM on two benchmark fMRI datasets: ABIDE and ADNI. Experimental results demonstrate that CP-SSM surpasses Transformer-based models in classification performance while significantly reducing computational complexity. These findings highlight the effectiveness and efficiency of CP-SSM in modeling brain functional connectivity, offering a promising direction for neuroimaging-based neurological disease diagnosis.
Brain-Adapter: Enhancing Neurological Disorder Analysis with Adapter-Tuning Multimodal Large Language Models
Jan 27, 2025


Abstract:Understanding brain disorders is crucial for accurate clinical diagnosis and treatment. Recent advances in Multimodal Large Language Models (MLLMs) offer a promising approach to interpreting medical images with the support of text descriptions. However, previous research has primarily focused on 2D medical images, leaving richer spatial information of 3D images under-explored, and single-modality-based methods are limited by overlooking the critical clinical information contained in other modalities. To address this issue, this paper proposes Brain-Adapter, a novel approach that incorporates an extra bottleneck layer to learn new knowledge and instill it into the original pre-trained knowledge. The major idea is to incorporate a lightweight bottleneck layer to train fewer parameters while capturing essential information and utilize a Contrastive Language-Image Pre-training (CLIP) strategy to align multimodal data within a unified representation space. Extensive experiments demonstrated the effectiveness of our approach in integrating multimodal data to significantly improve the diagnosis accuracy without high computational costs, highlighting the potential to enhance real-world diagnostic workflows.
Classification of Mild Cognitive Impairment Based on Dynamic Functional Connectivity Using Spatio-Temporal Transformer
Jan 27, 2025Abstract:Dynamic functional connectivity (dFC) using resting-state functional magnetic resonance imaging (rs-fMRI) is an advanced technique for capturing the dynamic changes of neural activities, and can be very useful in the studies of brain diseases such as Alzheimer's disease (AD). Yet, existing studies have not fully leveraged the sequential information embedded within dFC that can potentially provide valuable information when identifying brain conditions. In this paper, we propose a novel framework that jointly learns the embedding of both spatial and temporal information within dFC based on the transformer architecture. Specifically, we first construct dFC networks from rs-fMRI data through a sliding window strategy. Then, we simultaneously employ a temporal block and a spatial block to capture higher-order representations of dynamic spatio-temporal dependencies, via mapping them into an efficient fused feature representation. To further enhance the robustness of these feature representations by reducing the dependency on labeled data, we also introduce a contrastive learning strategy to manipulate different brain states. Experimental results on 345 subjects with 570 scans from the Alzheimer's Disease Neuroimaging Initiative (ADNI) demonstrate the superiority of our proposed method for MCI (Mild Cognitive Impairment, the prodromal stage of AD) prediction, highlighting its potential for early identification of AD.
Using Structural Similarity and Kolmogorov-Arnold Networks for Anatomical Embedding of 3-hinge Gyrus
Oct 31, 2024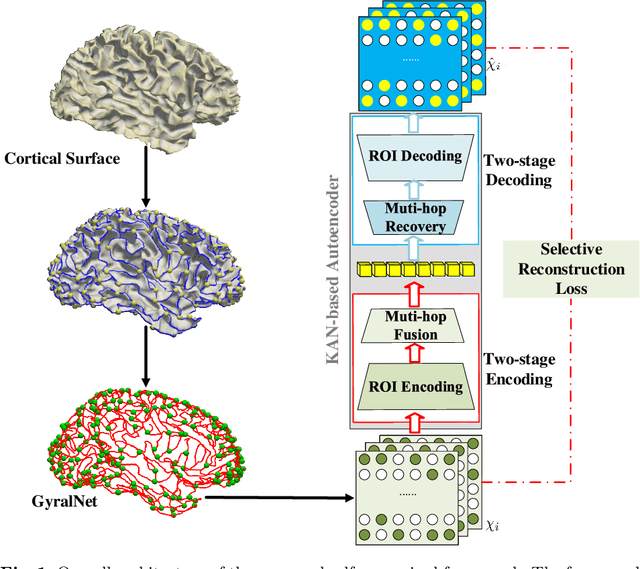
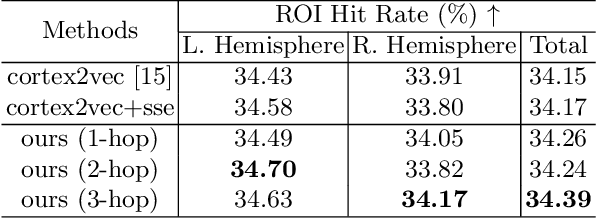
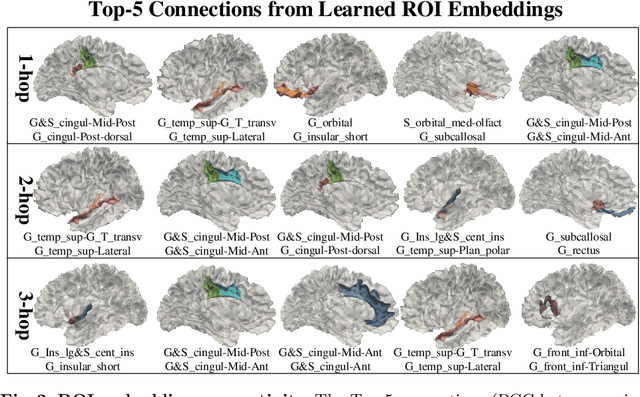

Abstract:The 3-hinge gyrus (3HG) is a newly defined folding pattern, which is the conjunction of gyri coming from three directions in cortical folding. Many studies demonstrated that 3HGs can be reliable nodes when constructing brain networks or connectome since they simultaneously possess commonality and individuality across different individual brains and populations. However, 3HGs are identified and validated within individual spaces, making it difficult to directly serve as the brain network nodes due to the absence of cross-subject correspondence. The 3HG correspondences represent the intrinsic regulation of brain organizational architecture, traditional image-based registration methods tend to fail because individual anatomical properties need to be fully respected. To address this challenge, we propose a novel self-supervised framework for anatomical feature embedding of the 3HGs to build the correspondences among different brains. The core component of this framework is to construct a structural similarity-enhanced multi-hop feature encoding strategy based on the recently developed Kolmogorov-Arnold network (KAN) for anatomical feature embedding. Extensive experiments suggest that our approach can effectively establish robust cross-subject correspondences when no one-to-one mapping exists.
Evaluation of OpenAI o1: Opportunities and Challenges of AGI
Sep 27, 2024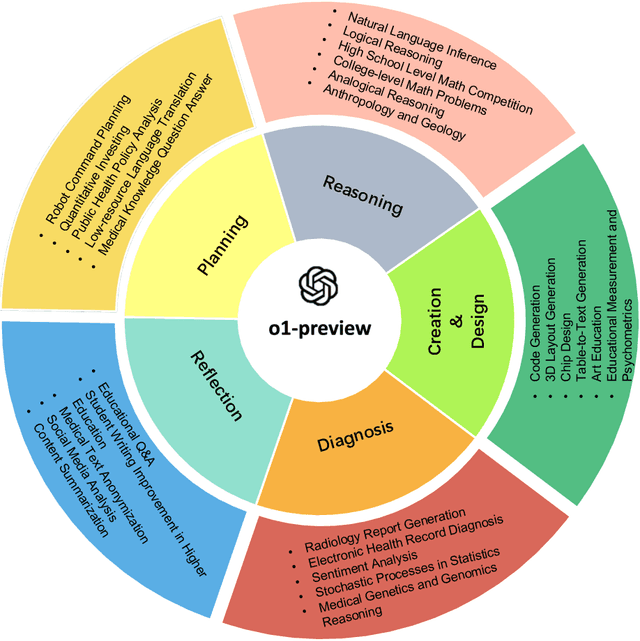
Abstract:This comprehensive study evaluates the performance of OpenAI's o1-preview large language model across a diverse array of complex reasoning tasks, spanning multiple domains, including computer science, mathematics, natural sciences, medicine, linguistics, and social sciences. Through rigorous testing, o1-preview demonstrated remarkable capabilities, often achieving human-level or superior performance in areas ranging from coding challenges to scientific reasoning and from language processing to creative problem-solving. Key findings include: -83.3% success rate in solving complex competitive programming problems, surpassing many human experts. -Superior ability in generating coherent and accurate radiology reports, outperforming other evaluated models. -100% accuracy in high school-level mathematical reasoning tasks, providing detailed step-by-step solutions. -Advanced natural language inference capabilities across general and specialized domains like medicine. -Impressive performance in chip design tasks, outperforming specialized models in areas such as EDA script generation and bug analysis. -Remarkable proficiency in anthropology and geology, demonstrating deep understanding and reasoning in these specialized fields. -Strong capabilities in quantitative investing. O1 has comprehensive financial knowledge and statistical modeling skills. -Effective performance in social media analysis, including sentiment analysis and emotion recognition. The model excelled particularly in tasks requiring intricate reasoning and knowledge integration across various fields. While some limitations were observed, including occasional errors on simpler problems and challenges with certain highly specialized concepts, the overall results indicate significant progress towards artificial general intelligence.
GP-GPT: Large Language Model for Gene-Phenotype Mapping
Sep 15, 2024



Abstract:Pre-trained large language models(LLMs) have attracted increasing attention in biomedical domains due to their success in natural language processing. However, the complex traits and heterogeneity of multi-sources genomics data pose significant challenges when adapting these models to the bioinformatics and biomedical field. To address these challenges, we present GP-GPT, the first specialized large language model for genetic-phenotype knowledge representation and genomics relation analysis. Our model is fine-tuned in two stages on a comprehensive corpus composed of over 3,000,000 terms in genomics, proteomics, and medical genetics, derived from multiple large-scale validated datasets and scientific publications. GP-GPT demonstrates proficiency in accurately retrieving medical genetics information and performing common genomics analysis tasks, such as genomics information retrieval and relationship determination. Comparative experiments across domain-specific tasks reveal that GP-GPT outperforms state-of-the-art LLMs, including Llama2, Llama3 and GPT-4. These results highlight GP-GPT's potential to enhance genetic disease relation research and facilitate accurate and efficient analysis in the fields of genomics and medical genetics. Our investigation demonstrated the subtle changes of bio-factor entities' representations in the GP-GPT, which suggested the opportunities for the application of LLMs to advancing gene-phenotype research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge