Xiaowei Yu
Learning Image-based Tree Crown Segmentation from Enhanced Lidar-based Pseudo-labels
Feb 13, 2026Abstract:Mapping individual tree crowns is essential for tasks such as maintaining urban tree inventories and monitoring forest health, which help us understand and care for our environment. However, automatically separating the crowns from each other in aerial imagery is challenging due to factors such as the texture and partial tree crown overlaps. In this study, we present a method to train deep learning models that segment and separate individual trees from RGB and multispectral images, using pseudo-labels derived from aerial laser scanning (ALS) data. Our study shows that the ALS-derived pseudo-labels can be enhanced using a zero-shot instance segmentation model, Segment Anything Model 2 (SAM 2). Our method offers a way to obtain domain-specific training annotations for optical image-based models without any manual annotation cost, leading to segmentation models which outperform any available models which have been targeted for general domain deployment on the same task.
DCMM-Transformer: Degree-Corrected Mixed-Membership Attention for Medical Imaging
Nov 15, 2025Abstract:Medical images exhibit latent anatomical groupings, such as organs, tissues, and pathological regions, that standard Vision Transformers (ViTs) fail to exploit. While recent work like SBM-Transformer attempts to incorporate such structures through stochastic binary masking, they suffer from non-differentiability, training instability, and the inability to model complex community structure. We present DCMM-Transformer, a novel ViT architecture for medical image analysis that incorporates a Degree-Corrected Mixed-Membership (DCMM) model as an additive bias in self-attention. Unlike prior approaches that rely on multiplicative masking and binary sampling, our method introduces community structure and degree heterogeneity in a fully differentiable and interpretable manner. Comprehensive experiments across diverse medical imaging datasets, including brain, chest, breast, and ocular modalities, demonstrate the superior performance and generalizability of the proposed approach. Furthermore, the learned group structure and structured attention modulation substantially enhance interpretability by yielding attention maps that are anatomically meaningful and semantically coherent.
Knowledge Distillation and Dataset Distillation of Large Language Models: Emerging Trends, Challenges, and Future Directions
Apr 20, 2025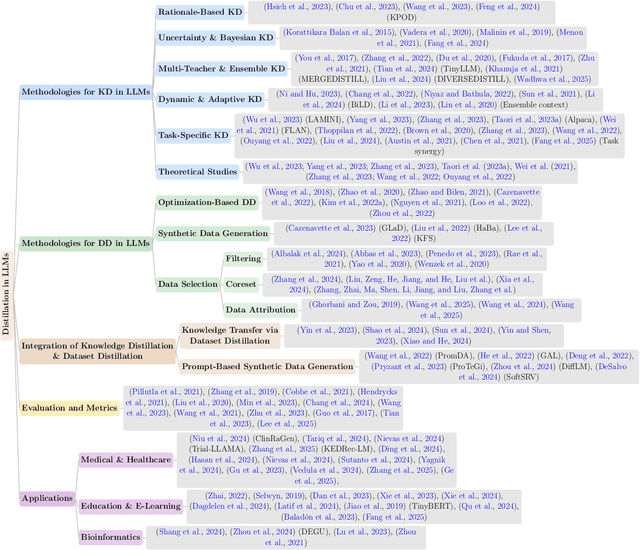
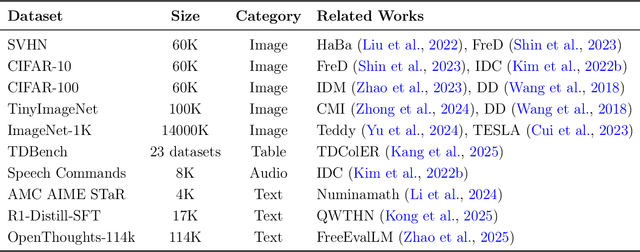
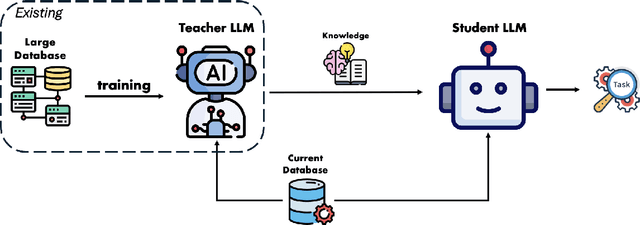
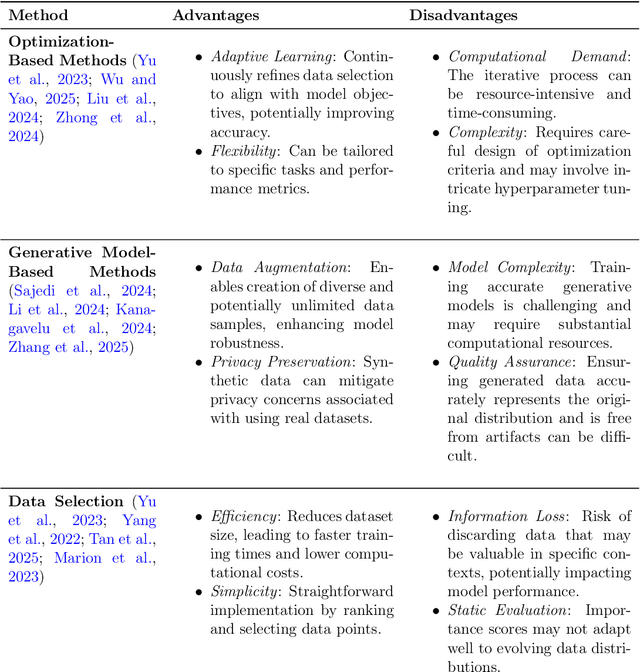
Abstract:The exponential growth of Large Language Models (LLMs) continues to highlight the need for efficient strategies to meet ever-expanding computational and data demands. This survey provides a comprehensive analysis of two complementary paradigms: Knowledge Distillation (KD) and Dataset Distillation (DD), both aimed at compressing LLMs while preserving their advanced reasoning capabilities and linguistic diversity. We first examine key methodologies in KD, such as task-specific alignment, rationale-based training, and multi-teacher frameworks, alongside DD techniques that synthesize compact, high-impact datasets through optimization-based gradient matching, latent space regularization, and generative synthesis. Building on these foundations, we explore how integrating KD and DD can produce more effective and scalable compression strategies. Together, these approaches address persistent challenges in model scalability, architectural heterogeneity, and the preservation of emergent LLM abilities. We further highlight applications across domains such as healthcare and education, where distillation enables efficient deployment without sacrificing performance. Despite substantial progress, open challenges remain in preserving emergent reasoning and linguistic diversity, enabling efficient adaptation to continually evolving teacher models and datasets, and establishing comprehensive evaluation protocols. By synthesizing methodological innovations, theoretical foundations, and practical insights, our survey charts a path toward sustainable, resource-efficient LLMs through the tighter integration of KD and DD principles.
Multispectral airborne laser scanning for tree species classification: a benchmark of machine learning and deep learning algorithms
Apr 19, 2025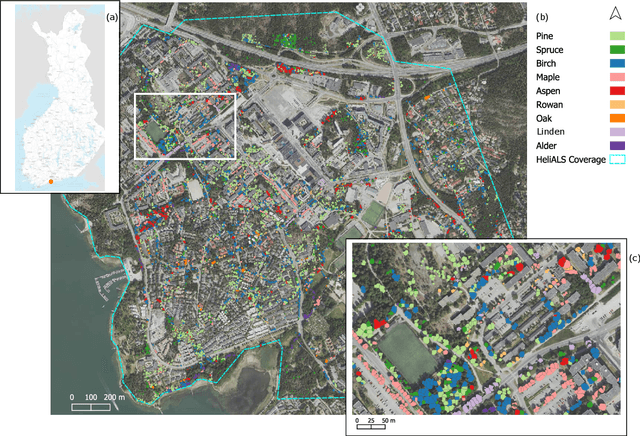
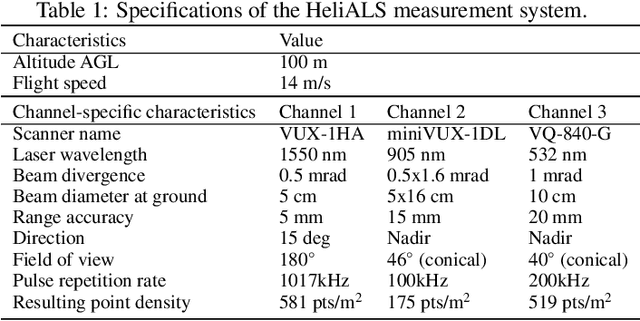

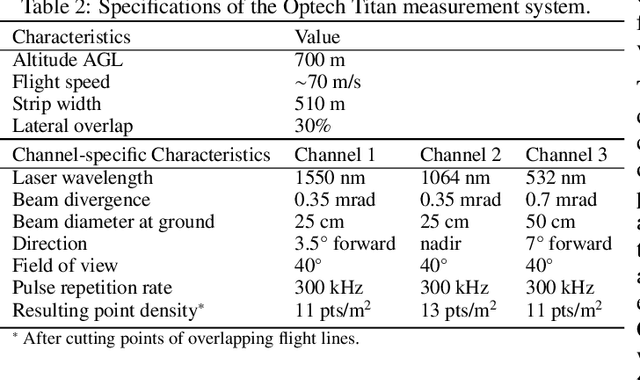
Abstract:Climate-smart and biodiversity-preserving forestry demands precise information on forest resources, extending to the individual tree level. Multispectral airborne laser scanning (ALS) has shown promise in automated point cloud processing and tree segmentation, but challenges remain in identifying rare tree species and leveraging deep learning techniques. This study addresses these gaps by conducting a comprehensive benchmark of machine learning and deep learning methods for tree species classification. For the study, we collected high-density multispectral ALS data (>1000 pts/m$^2$) at three wavelengths using the FGI-developed HeliALS system, complemented by existing Optech Titan data (35 pts/m$^2$), to evaluate the species classification accuracy of various algorithms in a test site located in Southern Finland. Based on 5261 test segments, our findings demonstrate that point-based deep learning methods, particularly a point transformer model, outperformed traditional machine learning and image-based deep learning approaches on high-density multispectral point clouds. For the high-density ALS dataset, a point transformer model provided the best performance reaching an overall (macro-average) accuracy of 87.9% (74.5%) with a training set of 1065 segments and 92.0% (85.1%) with 5000 training segments. The best image-based deep learning method, DetailView, reached an overall (macro-average) accuracy of 84.3% (63.9%), whereas a random forest (RF) classifier achieved an overall (macro-average) accuracy of 83.2% (61.3%). Importantly, the overall classification accuracy of the point transformer model on the HeliALS data increased from 73.0% with no spectral information to 84.7% with single-channel reflectance, and to 87.9% with spectral information of all the three channels.
GyralNet Subnetwork Partitioning via Differentiable Spectral Modularity Optimization
Mar 25, 2025



Abstract:Understanding the structural and functional organization of the human brain requires a detailed examination of cortical folding patterns, among which the three-hinge gyrus (3HG) has been identified as a key structural landmark. GyralNet, a network representation of cortical folding, models 3HGs as nodes and gyral crests as edges, highlighting their role as critical hubs in cortico-cortical connectivity. However, existing methods for analyzing 3HGs face significant challenges, including the sub-voxel scale of 3HGs at typical neuroimaging resolutions, the computational complexity of establishing cross-subject correspondences, and the oversimplification of treating 3HGs as independent nodes without considering their community-level relationships. To address these limitations, we propose a fully differentiable subnetwork partitioning framework that employs a spectral modularity maximization optimization strategy to modularize the organization of 3HGs within GyralNet. By incorporating topological structural similarity and DTI-derived connectivity patterns as attribute features, our approach provides a biologically meaningful representation of cortical organization. Extensive experiments on the Human Connectome Project (HCP) dataset demonstrate that our method effectively partitions GyralNet at the individual level while preserving the community-level consistency of 3HGs across subjects, offering a robust foundation for understanding brain connectivity.
Core-Periphery Principle Guided State Space Model for Functional Connectome Classification
Mar 18, 2025Abstract:Understanding the organization of human brain networks has become a central focus in neuroscience, particularly in the study of functional connectivity, which plays a crucial role in diagnosing neurological disorders. Advances in functional magnetic resonance imaging and machine learning techniques have significantly improved brain network analysis. However, traditional machine learning approaches struggle to capture the complex relationships between brain regions, while deep learning methods, particularly Transformer-based models, face computational challenges due to their quadratic complexity in long-sequence modeling. To address these limitations, we propose a Core-Periphery State-Space Model (CP-SSM), an innovative framework for functional connectome classification. Specifically, we introduce Mamba, a selective state-space model with linear complexity, to effectively capture long-range dependencies in functional brain networks. Furthermore, inspired by the core-periphery (CP) organization, a fundamental characteristic of brain networks that enhances efficient information transmission, we design CP-MoE, a CP-guided Mixture-of-Experts that improves the representation learning of brain connectivity patterns. We evaluate CP-SSM on two benchmark fMRI datasets: ABIDE and ADNI. Experimental results demonstrate that CP-SSM surpasses Transformer-based models in classification performance while significantly reducing computational complexity. These findings highlight the effectiveness and efficiency of CP-SSM in modeling brain functional connectivity, offering a promising direction for neuroimaging-based neurological disease diagnosis.
Brain-Adapter: Enhancing Neurological Disorder Analysis with Adapter-Tuning Multimodal Large Language Models
Jan 27, 2025


Abstract:Understanding brain disorders is crucial for accurate clinical diagnosis and treatment. Recent advances in Multimodal Large Language Models (MLLMs) offer a promising approach to interpreting medical images with the support of text descriptions. However, previous research has primarily focused on 2D medical images, leaving richer spatial information of 3D images under-explored, and single-modality-based methods are limited by overlooking the critical clinical information contained in other modalities. To address this issue, this paper proposes Brain-Adapter, a novel approach that incorporates an extra bottleneck layer to learn new knowledge and instill it into the original pre-trained knowledge. The major idea is to incorporate a lightweight bottleneck layer to train fewer parameters while capturing essential information and utilize a Contrastive Language-Image Pre-training (CLIP) strategy to align multimodal data within a unified representation space. Extensive experiments demonstrated the effectiveness of our approach in integrating multimodal data to significantly improve the diagnosis accuracy without high computational costs, highlighting the potential to enhance real-world diagnostic workflows.
Classification of Mild Cognitive Impairment Based on Dynamic Functional Connectivity Using Spatio-Temporal Transformer
Jan 27, 2025Abstract:Dynamic functional connectivity (dFC) using resting-state functional magnetic resonance imaging (rs-fMRI) is an advanced technique for capturing the dynamic changes of neural activities, and can be very useful in the studies of brain diseases such as Alzheimer's disease (AD). Yet, existing studies have not fully leveraged the sequential information embedded within dFC that can potentially provide valuable information when identifying brain conditions. In this paper, we propose a novel framework that jointly learns the embedding of both spatial and temporal information within dFC based on the transformer architecture. Specifically, we first construct dFC networks from rs-fMRI data through a sliding window strategy. Then, we simultaneously employ a temporal block and a spatial block to capture higher-order representations of dynamic spatio-temporal dependencies, via mapping them into an efficient fused feature representation. To further enhance the robustness of these feature representations by reducing the dependency on labeled data, we also introduce a contrastive learning strategy to manipulate different brain states. Experimental results on 345 subjects with 570 scans from the Alzheimer's Disease Neuroimaging Initiative (ADNI) demonstrate the superiority of our proposed method for MCI (Mild Cognitive Impairment, the prodromal stage of AD) prediction, highlighting its potential for early identification of AD.
Feature Fusion Transferability Aware Transformer for Unsupervised Domain Adaptation
Nov 10, 2024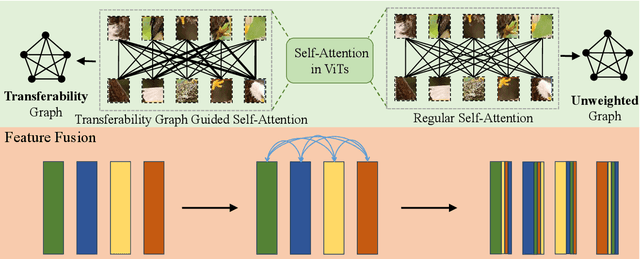
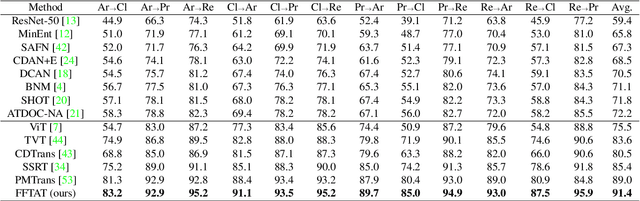
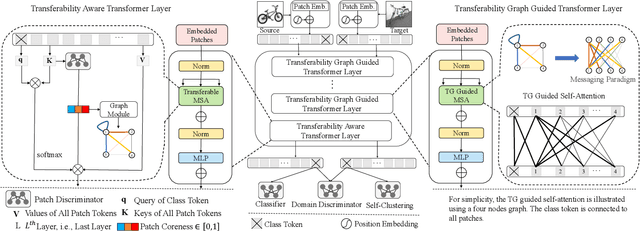
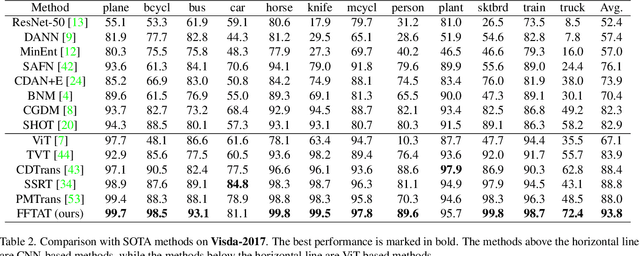
Abstract:Unsupervised domain adaptation (UDA) aims to leverage the knowledge learned from labeled source domains to improve performance on the unlabeled target domains. While Convolutional Neural Networks (CNNs) have been dominant in previous UDA methods, recent research has shown promise in applying Vision Transformers (ViTs) to this task. In this study, we propose a novel Feature Fusion Transferability Aware Transformer (FFTAT) to enhance ViT performance in UDA tasks. Our method introduces two key innovations: First, we introduce a patch discriminator to evaluate the transferability of patches, generating a transferability matrix. We integrate this matrix into self-attention, directing the model to focus on transferable patches. Second, we propose a feature fusion technique to fuse embeddings in the latent space, enabling each embedding to incorporate information from all others, thereby improving generalization. These two components work in synergy to enhance feature representation learning. Extensive experiments on widely used benchmarks demonstrate that our method significantly improves UDA performance, achieving state-of-the-art (SOTA) results.
Using Structural Similarity and Kolmogorov-Arnold Networks for Anatomical Embedding of 3-hinge Gyrus
Oct 31, 2024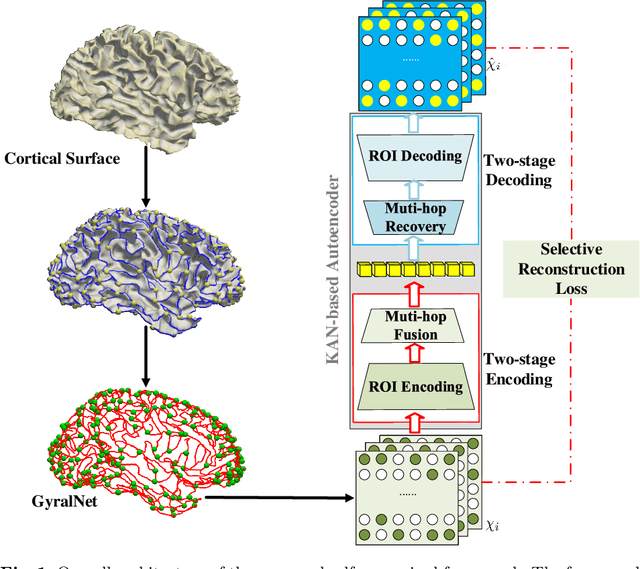
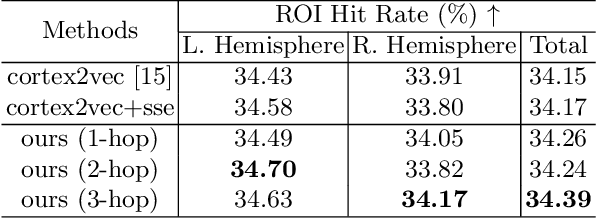
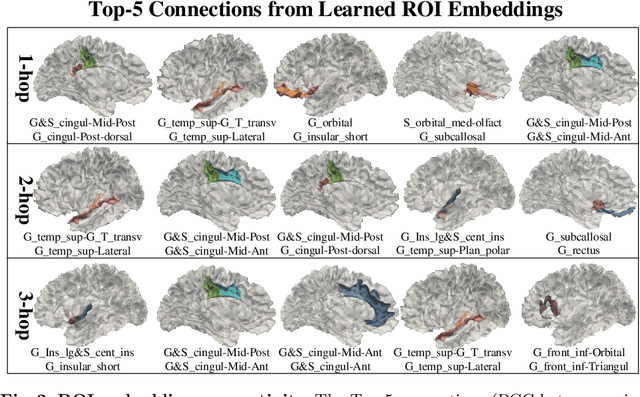

Abstract:The 3-hinge gyrus (3HG) is a newly defined folding pattern, which is the conjunction of gyri coming from three directions in cortical folding. Many studies demonstrated that 3HGs can be reliable nodes when constructing brain networks or connectome since they simultaneously possess commonality and individuality across different individual brains and populations. However, 3HGs are identified and validated within individual spaces, making it difficult to directly serve as the brain network nodes due to the absence of cross-subject correspondence. The 3HG correspondences represent the intrinsic regulation of brain organizational architecture, traditional image-based registration methods tend to fail because individual anatomical properties need to be fully respected. To address this challenge, we propose a novel self-supervised framework for anatomical feature embedding of the 3HGs to build the correspondences among different brains. The core component of this framework is to construct a structural similarity-enhanced multi-hop feature encoding strategy based on the recently developed Kolmogorov-Arnold network (KAN) for anatomical feature embedding. Extensive experiments suggest that our approach can effectively establish robust cross-subject correspondences when no one-to-one mapping exists.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge