Panagiotis Korfiatis
A Classification-Based Adaptive Segmentation Pipeline: Feasibility Study Using Polycystic Liver Disease and Metastases from Colorectal Cancer CT Images
May 02, 2024Abstract:Automated segmentation tools often encounter accuracy and adaptability issues when applied to images of different pathology. The purpose of this study is to explore the feasibility of building a workflow to efficiently route images to specifically trained segmentation models. By implementing a deep learning classifier to automatically classify the images and route them to appropriate segmentation models, we hope that our workflow can segment the images with different pathology accurately. The data we used in this study are 350 CT images from patients affected by polycystic liver disease and 350 CT images from patients presenting with liver metastases from colorectal cancer. All images had the liver manually segmented by trained imaging analysts. Our proposed adaptive segmentation workflow achieved a statistically significant improvement for the task of total liver segmentation compared to the generic single segmentation model (non-parametric Wilcoxon signed rank test, n=100, p-value << 0.001). This approach is applicable in a wide range of scenarios and should prove useful in clinical implementations of segmentation pipelines.
Role of Image Acquisition and Patient Phenotype Variations in Automatic Segmentation Model Generalization
Jul 26, 2023



Abstract:Purpose: This study evaluated the out-of-domain performance and generalization capabilities of automated medical image segmentation models, with a particular focus on adaptation to new image acquisitions and disease type. Materials: Datasets from both non-contrast and contrast-enhanced abdominal CT scans of healthy patients and those with polycystic kidney disease (PKD) were used. A total of 400 images (100 non-contrast controls, 100 contrast controls, 100 non-contrast PKD, 100 contrast PKD) were utilized for training/validation of models to segment kidneys, livers, and spleens, and the final models were then tested on 100 non-contrast CT images of patients affected by PKD. Performance was evaluated using Dice, Jaccard, TPR, and Precision. Results: Models trained on a diverse range of data showed no worse performance than models trained exclusively on in-domain data when tested on in-domain data. For instance, the Dice similarity of the model trained on 25% from each dataset was found to be non-inferior to the model trained purely on in-domain data. Conclusions: The results indicate that broader training examples significantly enhances model generalization and out-of-domain performance, thereby improving automated segmentation tools' applicability in clinical settings. The study's findings provide a roadmap for future research to adopt a data-centric approach in medical image AI model development.
AI in the Loop -- Functionalizing Fold Performance Disagreement to Monitor Automated Medical Image Segmentation Pipelines
May 15, 2023

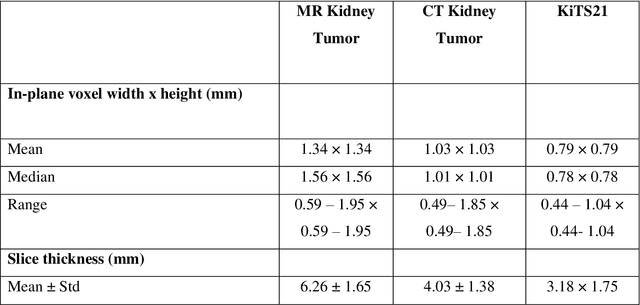

Abstract:Methods for automatically flag poor performing-predictions are essential for safely implementing machine learning workflows into clinical practice and for identifying difficult cases during model training. We present a readily adoptable method using sub-models trained on different dataset folds, where their disagreement serves as a surrogate for model confidence. Thresholds informed by human interobserver values were used to determine whether a final ensemble model prediction would require manual review. In two different datasets (abdominal CT and MR predicting kidney tumors), our framework effectively identified low performing automated segmentations. Flagging images with a minimum Interfold test Dice score below human interobserver variability maximized the number of flagged images while ensuring maximum ensemble test Dice. When our internally trained model was applied to an external publicly available dataset (KiTS21), flagged images included smaller tumors than those observed in our internally trained dataset, demonstrating the methods robustness to flagging poor performing out-of-distribution input data. Comparing interfold sub-model disagreement against human interobserver values is an efficient way to approximate a model's epistemic uncertainty - its lack of knowledge due to insufficient relevant training data - a key functionality for adopting these applications in clinical practice.
Reproducibility in medical image radiomic studies: contribution of dynamic histogram binning
Nov 09, 2022Abstract:The de facto standard of dynamic histogram binning for radiomic feature extraction leads to an elevated sensitivity to fluctuations in annotated regions. This may impact the majority of radiomic studies published recently and contribute to issues regarding poor reproducibility of radiomic-based machine learning that has led to significant efforts for data harmonization; however, we believe the issues highlighted here are comparatively neglected, but often remedied by choosing static binning. The field of radiomics has improved through the development of community standards and open-source libraries such as PyRadiomics. But differences in image acquisition, systematic differences between observers' annotations, and preprocessing steps still pose challenges. These can change the distribution of voxels altering extracted features and can be exacerbated with dynamic binning.
High Resolution Medical Image Analysis with Spatial Partitioning
Sep 12, 2019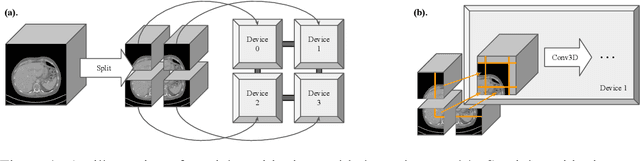
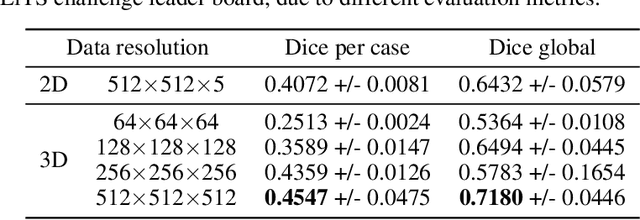
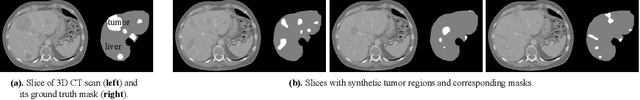
Abstract:Medical images such as 3D computerized tomography (CT) scans and pathology images, have hundreds of millions or billions of voxels/pixels. It is infeasible to train CNN models directly on such high resolution images, because neural activations of a single image do not fit in the memory of a single GPU/TPU, and naive data and model parallelism approaches do not work. Existing image analysis approaches alleviate this problem by cropping or down-sampling input images, which leads to complicated implementation and sub-optimal performance due to information loss. In this paper, we implement spatial partitioning, which internally distributes the input and output of convolutional layers across GPUs/TPUs. Our implementation is based on the Mesh-TensorFlow framework and the computation distribution is transparent to end users. With this technique, we train a 3D Unet on up to 512 by 512 by 512 resolution data. To the best of our knowledge, this is the first work for handling such high resolution images end-to-end.
Interactive segmentation of medical images through fully convolutional neural networks
Mar 19, 2019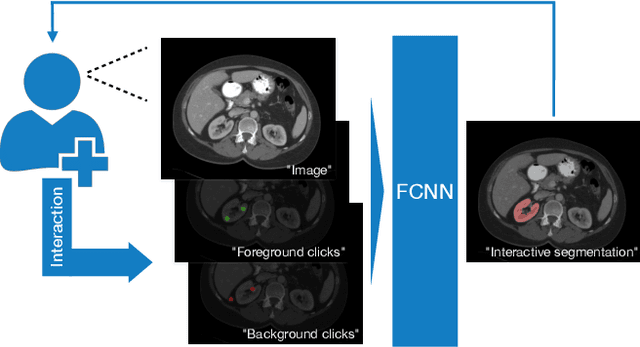



Abstract:Image segmentation plays an essential role in medicine for both diagnostic and interventional tasks. Segmentation approaches are either manual, semi-automated or fully-automated. Manual segmentation offers full control over the quality of the results, but is tedious, time consuming and prone to operator bias. Fully automated methods require no human effort, but often deliver sub-optimal results without providing users with the means to make corrections. Semi-automated approaches keep users in control of the results by providing means for interaction, but the main challenge is to offer a good trade-off between precision and required interaction. In this paper we present a deep learning (DL) based semi-automated segmentation approach that aims to be a "smart" interactive tool for region of interest delineation in medical images. We demonstrate its use for segmenting multiple organs on computed tomography (CT) of the abdomen. Our approach solves some of the most pressing clinical challenges: (i) it requires only one to a few user clicks to deliver excellent 2D segmentations in a fast and reliable fashion; (ii) it can generalize to previously unseen structures and "corner cases"; (iii) it delivers results that can be corrected quickly in a smart and intuitive way up to an arbitrary degree of precision chosen by the user and (iv) ensures high accuracy. We present our approach and compare it to other techniques and previous work to show the advantages brought by our method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge