Zeynettin Akkus
Enhancing Thyroid Cytology Diagnosis with RAG-Optimized LLMs and Pa-thology Foundation Models
May 13, 2025Abstract:Advancements in artificial intelligence (AI) are transforming pathology by integrat-ing large language models (LLMs) with retrieval-augmented generation (RAG) and domain-specific foundation models. This study explores the application of RAG-enhanced LLMs coupled with pathology foundation models for thyroid cytology diagnosis, addressing challenges in cytological interpretation, standardization, and diagnostic accuracy. By leveraging a curated knowledge base, RAG facilitates dy-namic retrieval of relevant case studies, diagnostic criteria, and expert interpreta-tion, improving the contextual understanding of LLMs. Meanwhile, pathology foun-dation models, trained on high-resolution pathology images, refine feature extrac-tion and classification capabilities. The fusion of these AI-driven approaches en-hances diagnostic consistency, reduces variability, and supports pathologists in dis-tinguishing benign from malignant thyroid lesions. Our results demonstrate that integrating RAG with pathology-specific LLMs significantly improves diagnostic efficiency and interpretability, paving the way for AI-assisted thyroid cytopathology, with foundation model UNI achieving AUC 0.73-0.93 for correct prediction of surgi-cal pathology diagnosis from thyroid cytology samples.
Fully Automated Mitral Inflow Doppler Analysis Using Deep Learning
Nov 24, 2020



Abstract:Echocardiography (echo) is an indispensable tool in a cardiologist's diagnostic armamentarium. To date, almost all echocardiographic parameters require time-consuming manual labeling and measurements by an experienced echocardiographer and exhibit significant variability, owing to the noisy and artifact-laden nature of echo images. For example, mitral inflow (MI) Doppler is used to assess left ventricular (LV) diastolic function, which is of paramount clinical importance to distinguish between different cardiac diseases. In the current work we present a fully automated workflow which leverages deep learning to a) label MI Doppler images acquired in an echo study, b) detect the envelope of MI Doppler signal, c) extract early and late filing (E and A wave) flow velocities and E-wave deceleration time from the envelope. We trained a variety of convolutional neural networks (CNN) models on 5544 images of 140 patients for predicting 24 image classes including MI Doppler images and obtained overall accuracy of 0.97 on 1737 images of 40 patients. Automated E and A wave velocity showed excellent correlation (Pearson R 0.99 and 0.98 respectively) and Bland Altman agreement (mean difference 0.06 and 0.05 m/s respectively and SD 0.03 for both) with the operator measurements. Deceleration time also showed good but lower correlation (Pearson R 0.82) and Bland-Altman agreement (mean difference: 34.1ms, SD: 30.9ms). These results demonstrate feasibility of Doppler echocardiography measurement automation and the promise of a fully automated echocardiography measurement package.
Interactive segmentation of medical images through fully convolutional neural networks
Mar 19, 2019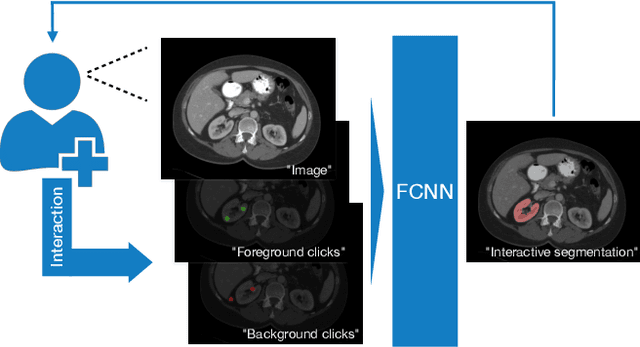



Abstract:Image segmentation plays an essential role in medicine for both diagnostic and interventional tasks. Segmentation approaches are either manual, semi-automated or fully-automated. Manual segmentation offers full control over the quality of the results, but is tedious, time consuming and prone to operator bias. Fully automated methods require no human effort, but often deliver sub-optimal results without providing users with the means to make corrections. Semi-automated approaches keep users in control of the results by providing means for interaction, but the main challenge is to offer a good trade-off between precision and required interaction. In this paper we present a deep learning (DL) based semi-automated segmentation approach that aims to be a "smart" interactive tool for region of interest delineation in medical images. We demonstrate its use for segmenting multiple organs on computed tomography (CT) of the abdomen. Our approach solves some of the most pressing clinical challenges: (i) it requires only one to a few user clicks to deliver excellent 2D segmentations in a fast and reliable fashion; (ii) it can generalize to previously unseen structures and "corner cases"; (iii) it delivers results that can be corrected quickly in a smart and intuitive way up to an arbitrary degree of precision chosen by the user and (iv) ensures high accuracy. We present our approach and compare it to other techniques and previous work to show the advantages brought by our method.
Predicting 1p19q Chromosomal Deletion of Low-Grade Gliomas from MR Images using Deep Learning
Nov 21, 2016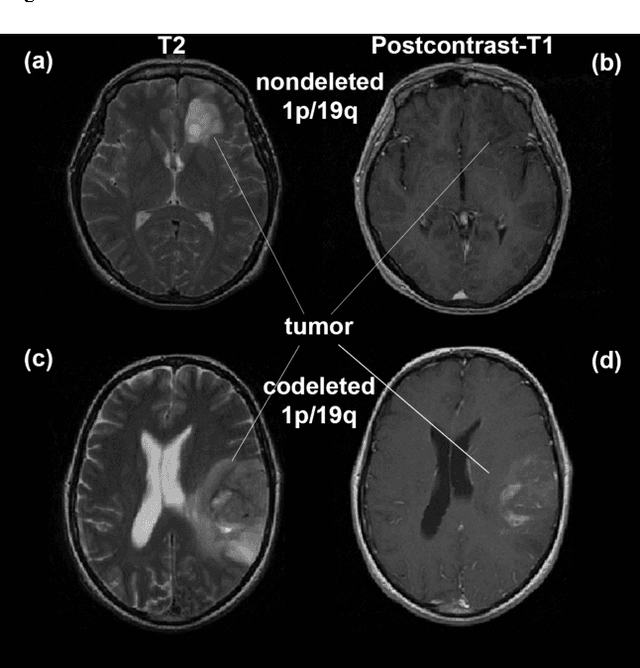
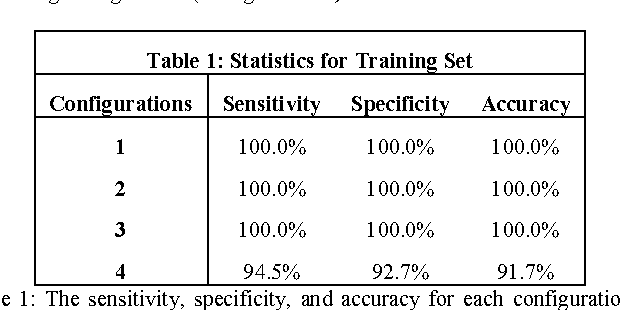


Abstract:Objective: Several studies have associated codeletion of chromosome arms 1p/19q in low-grade gliomas (LGG) with positive response to treatment and longer progression free survival. Therefore, predicting 1p/19q status is crucial for effective treatment planning of LGG. In this study, we predict the 1p/19q status from MR images using convolutional neural networks (CNN), which could be a noninvasive alternative to surgical biopsy and histopathological analysis. Method: Our method consists of three main steps: image registration, tumor segmentation, and classification of 1p/19q status using CNN. We included a total of 159 LGG with 3 image slices each who had biopsy-proven 1p/19q status (57 nondeleted and 102 codeleted) and preoperative postcontrast-T1 (T1C) and T2 images. We divided our data into training, validation, and test sets. The training data was balanced for equal class probability and then augmented with iterations of random translational shift, rotation, and horizontal and vertical flips to increase the size of the training set. We shuffled and augmented the training data to counter overfitting in each epoch. Finally, we evaluated several configurations of a multi-scale CNN architecture until training and validation accuracies became consistent. Results: The results of the best performing configuration on the unseen test set were 93.3% (sensitivity), 82.22% (specificity), and 87.7% (accuracy). Conclusion: Multi-scale CNN with their self-learning capability provides promising results for predicting 1p/19q status noninvasively based on T1C and T2 images. Significance: Predicting 1p/19q status noninvasively from MR images would allow selecting effective treatment strategies for LGG patients without the need for surgical biopsy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge