Predicting 1p19q Chromosomal Deletion of Low-Grade Gliomas from MR Images using Deep Learning
Paper and Code
Nov 21, 2016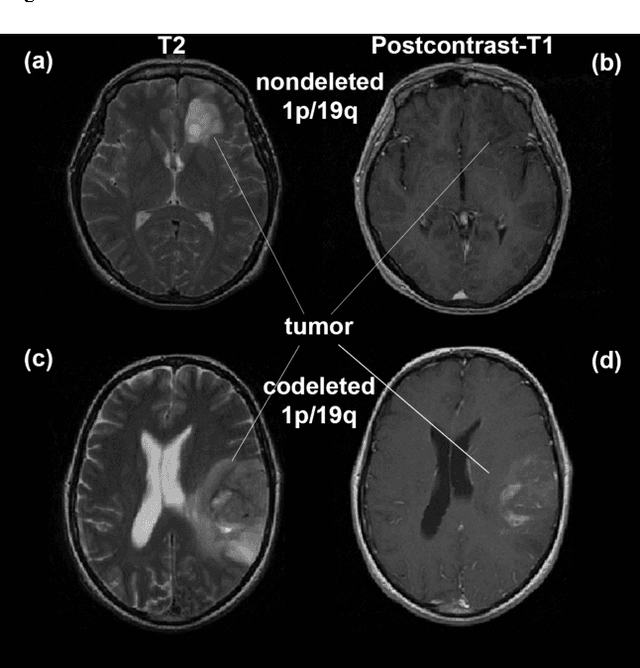
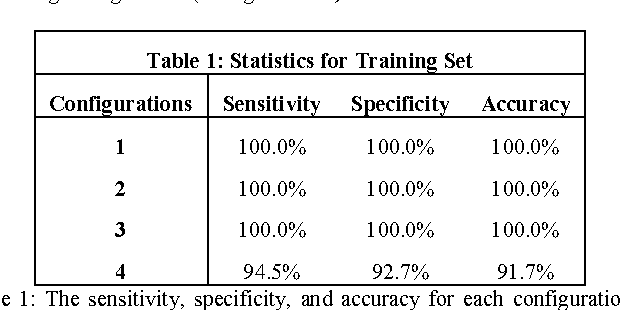


Objective: Several studies have associated codeletion of chromosome arms 1p/19q in low-grade gliomas (LGG) with positive response to treatment and longer progression free survival. Therefore, predicting 1p/19q status is crucial for effective treatment planning of LGG. In this study, we predict the 1p/19q status from MR images using convolutional neural networks (CNN), which could be a noninvasive alternative to surgical biopsy and histopathological analysis. Method: Our method consists of three main steps: image registration, tumor segmentation, and classification of 1p/19q status using CNN. We included a total of 159 LGG with 3 image slices each who had biopsy-proven 1p/19q status (57 nondeleted and 102 codeleted) and preoperative postcontrast-T1 (T1C) and T2 images. We divided our data into training, validation, and test sets. The training data was balanced for equal class probability and then augmented with iterations of random translational shift, rotation, and horizontal and vertical flips to increase the size of the training set. We shuffled and augmented the training data to counter overfitting in each epoch. Finally, we evaluated several configurations of a multi-scale CNN architecture until training and validation accuracies became consistent. Results: The results of the best performing configuration on the unseen test set were 93.3% (sensitivity), 82.22% (specificity), and 87.7% (accuracy). Conclusion: Multi-scale CNN with their self-learning capability provides promising results for predicting 1p/19q status noninvasively based on T1C and T2 images. Significance: Predicting 1p/19q status noninvasively from MR images would allow selecting effective treatment strategies for LGG patients without the need for surgical biopsy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge