Bradley J. Erickson
HOPPR Medical-Grade Platform for Medical Imaging AI
Nov 26, 2024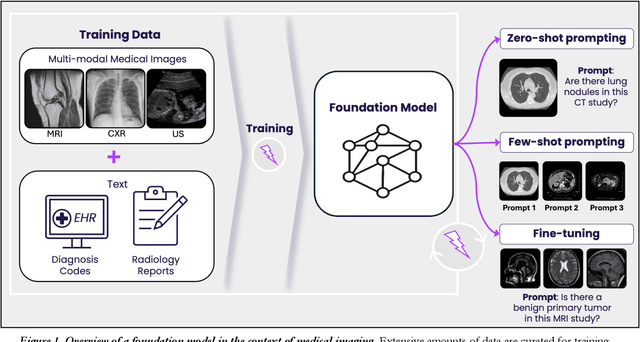
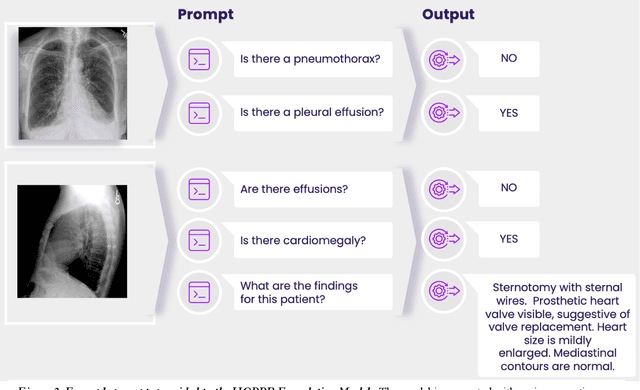
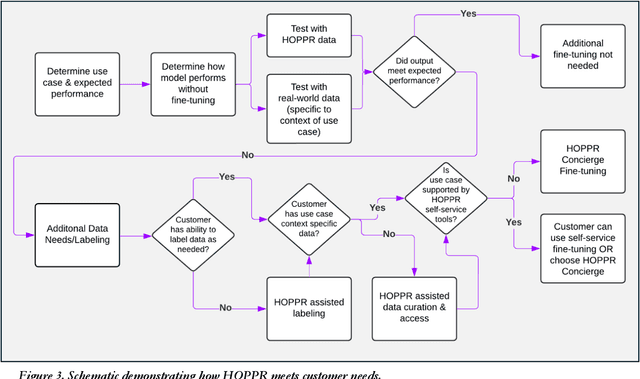
Abstract:Technological advances in artificial intelligence (AI) have enabled the development of large vision language models (LVLMs) that are trained on millions of paired image and text samples. Subsequent research efforts have demonstrated great potential of LVLMs to achieve high performance in medical imaging use cases (e.g., radiology report generation), but there remain barriers that hinder the ability to deploy these solutions broadly. These include the cost of extensive computational requirements for developing large scale models, expertise in the development of sophisticated AI models, and the difficulty in accessing substantially large, high-quality datasets that adequately represent the population in which the LVLM solution is to be deployed. The HOPPR Medical-Grade Platform addresses these barriers by providing powerful computational infrastructure, a suite of foundation models on top of which developers can fine-tune for their specific use cases, and a robust quality management system that sets a standard for evaluating fine-tuned models for deployment in clinical settings. The HOPPR Platform has access to millions of imaging studies and text reports sourced from hundreds of imaging centers from diverse populations to pretrain foundation models and enable use case-specific cohorts for fine-tuning. All data are deidentified and securely stored for HIPAA compliance. Additionally, developers can securely host models on the HOPPR platform and access them via an API to make inferences using these models within established clinical workflows. With the Medical-Grade Platform, HOPPR's mission is to expedite the deployment of LVLM solutions for medical imaging and ultimately optimize radiologist's workflows and meet the growing demands of the field.
RadRotator: 3D Rotation of Radiographs with Diffusion Models
Apr 19, 2024Abstract:Transforming two-dimensional (2D) images into three-dimensional (3D) volumes is a well-known yet challenging problem for the computer vision community. In the medical domain, a few previous studies attempted to convert two or more input radiographs into computed tomography (CT) volumes. Following their effort, we introduce a diffusion model-based technology that can rotate the anatomical content of any input radiograph in 3D space, potentially enabling the visualization of the entire anatomical content of the radiograph from any viewpoint in 3D. Similar to previous studies, we used CT volumes to create Digitally Reconstructed Radiographs (DRRs) as the training data for our model. However, we addressed two significant limitations encountered in previous studies: 1. We utilized conditional diffusion models with classifier-free guidance instead of Generative Adversarial Networks (GANs) to achieve higher mode coverage and improved output image quality, with the only trade-off being slower inference time, which is often less critical in medical applications; and 2. We demonstrated that the unreliable output of style transfer deep learning (DL) models, such as Cycle-GAN, to transfer the style of actual radiographs to DRRs could be replaced with a simple yet effective training transformation that randomly changes the pixel intensity histograms of the input and ground-truth imaging data during training. This transformation makes the diffusion model agnostic to any distribution variations of the input data pixel intensity, enabling the reliable training of a DL model on input DRRs and applying the exact same model to conventional radiographs (or DRRs) during inference.
CONFLARE: CONFormal LArge language model REtrieval
Apr 04, 2024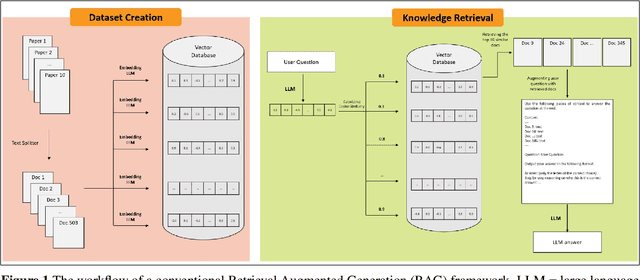
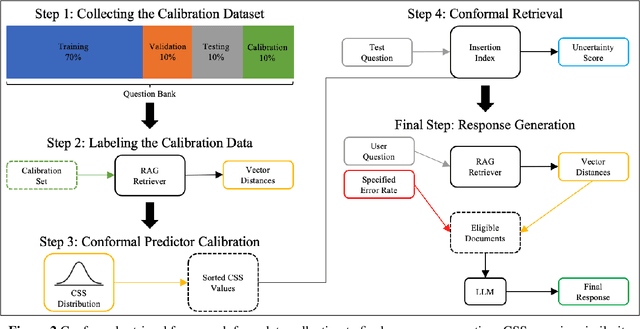
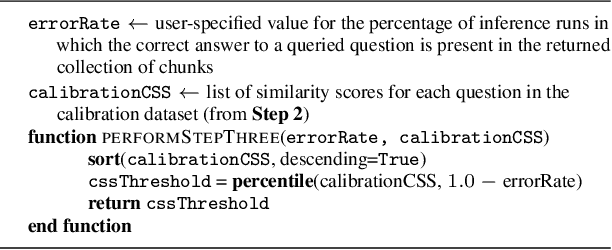

Abstract:Retrieval-augmented generation (RAG) frameworks enable large language models (LLMs) to retrieve relevant information from a knowledge base and incorporate it into the context for generating responses. This mitigates hallucinations and allows for the updating of knowledge without retraining the LLM. However, RAG does not guarantee valid responses if retrieval fails to identify the necessary information as the context for response generation. Also, if there is contradictory content, the RAG response will likely reflect only one of the two possible responses. Therefore, quantifying uncertainty in the retrieval process is crucial for ensuring RAG trustworthiness. In this report, we introduce a four-step framework for applying conformal prediction to quantify retrieval uncertainty in RAG frameworks. First, a calibration set of questions answerable from the knowledge base is constructed. Each question's embedding is compared against document embeddings to identify the most relevant document chunks containing the answer and record their similarity scores. Given a user-specified error rate ({\alpha}), these similarity scores are then analyzed to determine a similarity score cutoff threshold. During inference, all chunks with similarity exceeding this threshold are retrieved to provide context to the LLM, ensuring the true answer is captured in the context with a (1-{\alpha}) confidence level. We provide a Python package that enables users to implement the entire workflow proposed in our work, only using LLMs and without human intervention.
Toward Clinically Trustworthy Deep Learning: Applying Conformal Prediction to Intracranial Hemorrhage Detection
Jan 16, 2024Abstract:As deep learning (DL) continues to demonstrate its ability in radiological tasks, it is critical that we optimize clinical DL solutions to include safety. One of the principal concerns in the clinical adoption of DL tools is trust. This study aims to apply conformal prediction as a step toward trustworthiness for DL in radiology. This is a retrospective study of 491 non-contrast head CTs from the CQ500 dataset, in which three senior radiologists annotated slices containing intracranial hemorrhage (ICH). The dataset was split into definite and challenging subsets, where challenging images were defined to those in which there was disagreement among readers. A DL model was trained on 146 patients (10,815 slices) from the definite data (training dataset) to perform ICH localization and classification for five classes of ICH. To develop an uncertainty-aware DL model, 1,546 cases of the definite data (calibration dataset) was used for Mondrian conformal prediction (MCP). The uncertainty-aware DL model was tested on 8,401 definite and challenging cases to assess its ability to identify challenging cases. After the MCP procedure, the model achieved an F1 score of 0.920 for ICH classification on the test dataset. Additionally, it correctly identified 6,837 of the 6,856 total challenging cases as challenging (99.7% accuracy). It did not incorrectly label any definite cases as challenging. The uncertainty-aware ICH detector performs on par with state-of-the-art models. MCP's performance in detecting challenging cases demonstrates that it is useful in automated ICH detection and promising for trustworthiness in radiological DL.
MedYOLO: A Medical Image Object Detection Framework
Dec 12, 2023Abstract:Artificial intelligence-enhanced identification of organs, lesions, and other structures in medical imaging is typically done using convolutional neural networks (CNNs) designed to make voxel-accurate segmentations of the region of interest. However, the labels required to train these CNNs are time-consuming to generate and require attention from subject matter experts to ensure quality. For tasks where voxel-level precision is not required, object detection models offer a viable alternative that can reduce annotation effort. Despite this potential application, there are few options for general purpose object detection frameworks available for 3-D medical imaging. We report on MedYOLO, a 3-D object detection framework using the one-shot detection method of the YOLO family of models and designed for use with medical imaging. We tested this model on four different datasets: BRaTS, LIDC, an abdominal organ Computed Tomography (CT) dataset, and an ECG-gated heart CT dataset. We found our models achieve high performance on commonly present medium and large-sized structures such as the heart, liver, and pancreas even without hyperparameter tuning. However, the models struggle with very small or rarely present structures.
Synthetically Enhanced: Unveiling Synthetic Data's Potential in Medical Imaging Research
Nov 15, 2023



Abstract:Chest X-rays (CXR) are the most common medical imaging study and are used to diagnose multiple medical conditions. This study examines the impact of synthetic data supplementation, using diffusion models, on the performance of deep learning (DL) classifiers for CXR analysis. We employed three datasets: CheXpert, MIMIC-CXR, and Emory Chest X-ray, training conditional denoising diffusion probabilistic models (DDPMs) to generate synthetic frontal radiographs. Our approach ensured that synthetic images mirrored the demographic and pathological traits of the original data. Evaluating the classifiers' performance on internal and external datasets revealed that synthetic data supplementation enhances model accuracy, particularly in detecting less prevalent pathologies. Furthermore, models trained on synthetic data alone approached the performance of those trained on real data. This suggests that synthetic data can potentially compensate for real data shortages in training robust DL models. However, despite promising outcomes, the superiority of real data persists.
SOUP-GAN: Super-Resolution MRI Using Generative Adversarial Networks
Jun 04, 2021
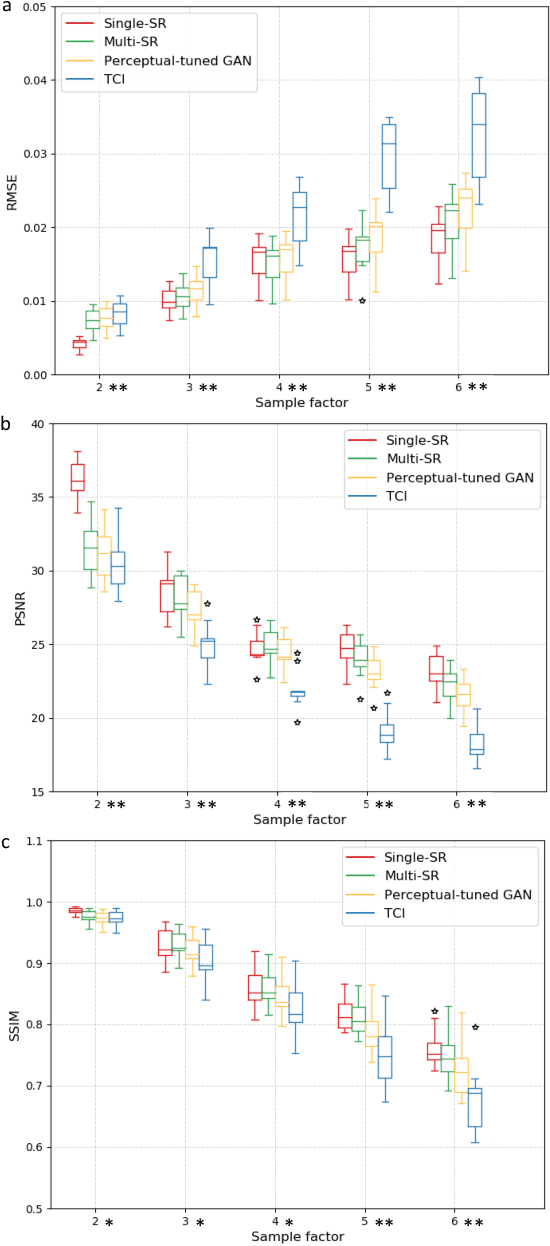
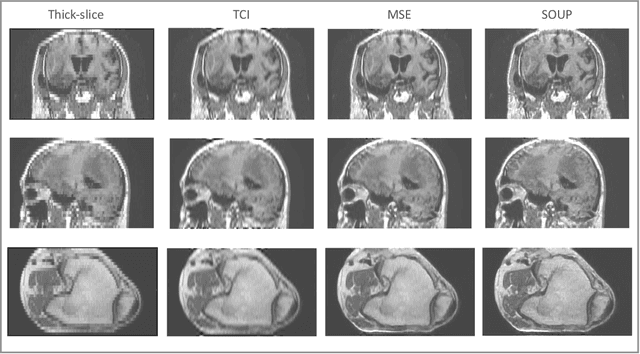
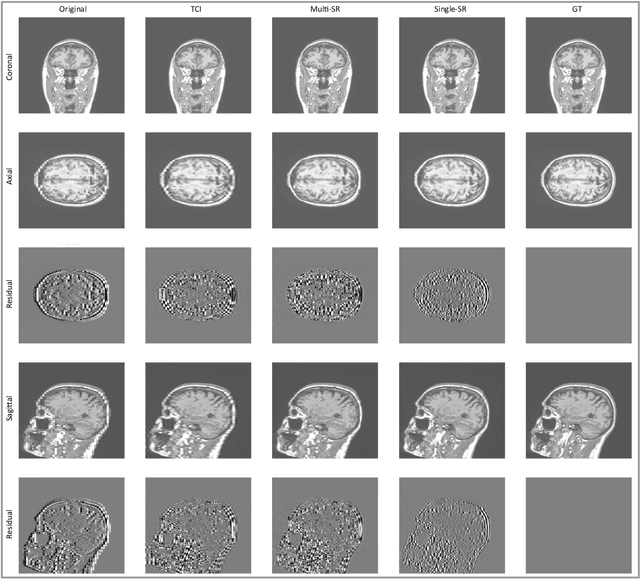
Abstract:There is a growing demand for high-resolution (HR) medical images in both the clinical and research applications. Image quality is inevitably traded off with the acquisition time for better patient comfort, lower examination costs, dose, and fewer motion-induced artifacts. For many image-based tasks, increasing the apparent resolution in the perpendicular plane to produce multi-planar reformats or 3D images is commonly used. Single image super-resolution (SR) is a promising technique to provide HR images based on unsupervised learning to increase resolution of a 2D image, but there are few reports on 3D SR. Further, perceptual loss is proposed in the literature to better capture the textual details and edges than using pixel-wise loss functions, by comparing the semantic distances in the high-dimensional feature space of a pre-trained 2D network (e.g., VGG). However, it is not clear how one should generalize it to 3D medical images, and the attendant implications are still unclear. In this paper, we propose a framework called SOUP-GAN: Super-resolution Optimized Using Perceptual-tuned Generative Adversarial Network (GAN), in order to produce thinner slice (e.g., high resolution in the 'Z' plane) medical images with anti-aliasing and deblurring. The proposed method outperforms other conventional resolution-enhancement methods and previous SR work on medical images upon both qualitative and quantitative comparisons. Specifically, we examine the model in terms of its generalization for various SR ratios and imaging modalities. By addressing those limitations, our model shows promise as a novel 3D SR interpolation technique, providing potential applications in both clinical and research settings.
Interactive segmentation of medical images through fully convolutional neural networks
Mar 19, 2019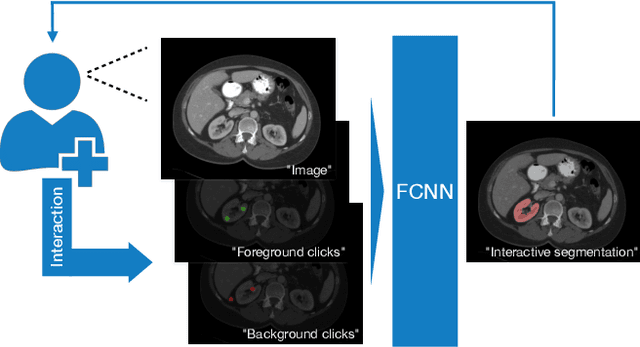



Abstract:Image segmentation plays an essential role in medicine for both diagnostic and interventional tasks. Segmentation approaches are either manual, semi-automated or fully-automated. Manual segmentation offers full control over the quality of the results, but is tedious, time consuming and prone to operator bias. Fully automated methods require no human effort, but often deliver sub-optimal results without providing users with the means to make corrections. Semi-automated approaches keep users in control of the results by providing means for interaction, but the main challenge is to offer a good trade-off between precision and required interaction. In this paper we present a deep learning (DL) based semi-automated segmentation approach that aims to be a "smart" interactive tool for region of interest delineation in medical images. We demonstrate its use for segmenting multiple organs on computed tomography (CT) of the abdomen. Our approach solves some of the most pressing clinical challenges: (i) it requires only one to a few user clicks to deliver excellent 2D segmentations in a fast and reliable fashion; (ii) it can generalize to previously unseen structures and "corner cases"; (iii) it delivers results that can be corrected quickly in a smart and intuitive way up to an arbitrary degree of precision chosen by the user and (iv) ensures high accuracy. We present our approach and compare it to other techniques and previous work to show the advantages brought by our method.
Predicting 1p19q Chromosomal Deletion of Low-Grade Gliomas from MR Images using Deep Learning
Nov 21, 2016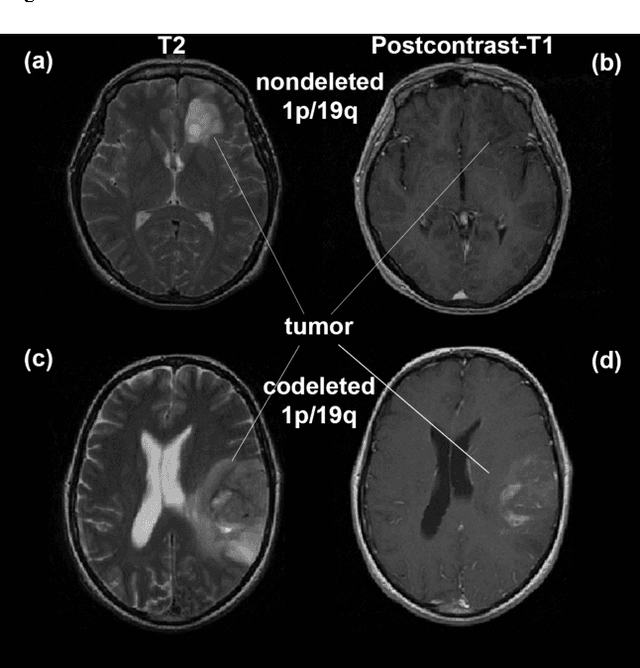
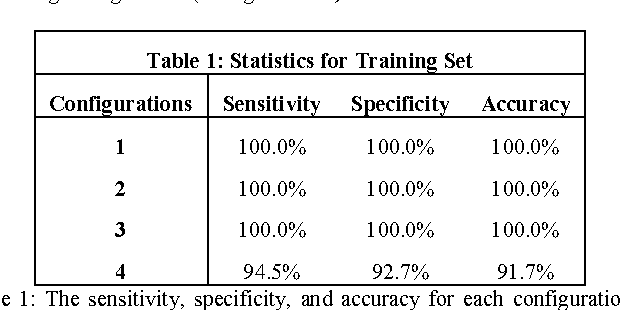


Abstract:Objective: Several studies have associated codeletion of chromosome arms 1p/19q in low-grade gliomas (LGG) with positive response to treatment and longer progression free survival. Therefore, predicting 1p/19q status is crucial for effective treatment planning of LGG. In this study, we predict the 1p/19q status from MR images using convolutional neural networks (CNN), which could be a noninvasive alternative to surgical biopsy and histopathological analysis. Method: Our method consists of three main steps: image registration, tumor segmentation, and classification of 1p/19q status using CNN. We included a total of 159 LGG with 3 image slices each who had biopsy-proven 1p/19q status (57 nondeleted and 102 codeleted) and preoperative postcontrast-T1 (T1C) and T2 images. We divided our data into training, validation, and test sets. The training data was balanced for equal class probability and then augmented with iterations of random translational shift, rotation, and horizontal and vertical flips to increase the size of the training set. We shuffled and augmented the training data to counter overfitting in each epoch. Finally, we evaluated several configurations of a multi-scale CNN architecture until training and validation accuracies became consistent. Results: The results of the best performing configuration on the unseen test set were 93.3% (sensitivity), 82.22% (specificity), and 87.7% (accuracy). Conclusion: Multi-scale CNN with their self-learning capability provides promising results for predicting 1p/19q status noninvasively based on T1C and T2 images. Significance: Predicting 1p/19q status noninvasively from MR images would allow selecting effective treatment strategies for LGG patients without the need for surgical biopsy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge