Gian Marco Conte
Analysis of the MICCAI Brain Tumor Segmentation -- Metastases (BraTS-METS) 2025 Lighthouse Challenge: Brain Metastasis Segmentation on Pre- and Post-treatment MRI
Apr 16, 2025Abstract:Despite continuous advancements in cancer treatment, brain metastatic disease remains a significant complication of primary cancer and is associated with an unfavorable prognosis. One approach for improving diagnosis, management, and outcomes is to implement algorithms based on artificial intelligence for the automated segmentation of both pre- and post-treatment MRI brain images. Such algorithms rely on volumetric criteria for lesion identification and treatment response assessment, which are still not available in clinical practice. Therefore, it is critical to establish tools for rapid volumetric segmentations methods that can be translated to clinical practice and that are trained on high quality annotated data. The BraTS-METS 2025 Lighthouse Challenge aims to address this critical need by establishing inter-rater and intra-rater variability in dataset annotation by generating high quality annotated datasets from four individual instances of segmentation by neuroradiologists while being recorded on video (two instances doing "from scratch" and two instances after AI pre-segmentation). This high-quality annotated dataset will be used for testing phase in 2025 Lighthouse challenge and will be publicly released at the completion of the challenge. The 2025 Lighthouse challenge will also release the 2023 and 2024 segmented datasets that were annotated using an established pipeline of pre-segmentation, student annotation, two neuroradiologists checking, and one neuroradiologist finalizing the process. It builds upon its previous edition by including post-treatment cases in the dataset. Using these high-quality annotated datasets, the 2025 Lighthouse challenge plans to test benchmark algorithms for automated segmentation of pre-and post-treatment brain metastases (BM), trained on diverse and multi-institutional datasets of MRI images obtained from patients with brain metastases.
The 2024 Brain Tumor Segmentation (BraTS) Challenge: Glioma Segmentation on Post-treatment MRI
May 28, 2024
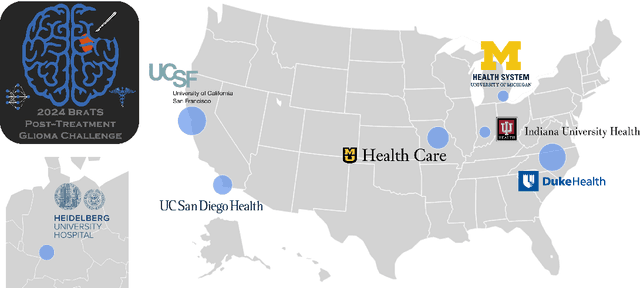
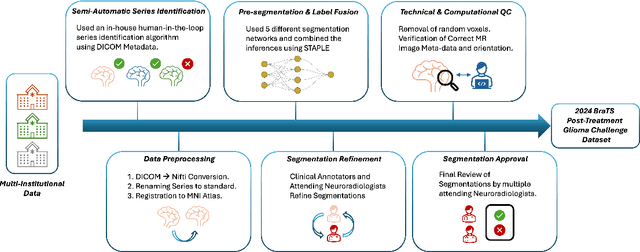
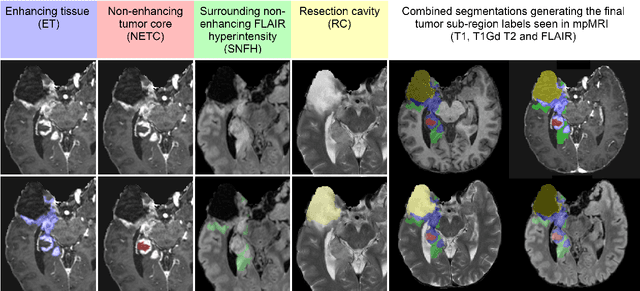
Abstract:Gliomas are the most common malignant primary brain tumors in adults and one of the deadliest types of cancer. There are many challenges in treatment and monitoring due to the genetic diversity and high intrinsic heterogeneity in appearance, shape, histology, and treatment response. Treatments include surgery, radiation, and systemic therapies, with magnetic resonance imaging (MRI) playing a key role in treatment planning and post-treatment longitudinal assessment. The 2024 Brain Tumor Segmentation (BraTS) challenge on post-treatment glioma MRI will provide a community standard and benchmark for state-of-the-art automated segmentation models based on the largest expert-annotated post-treatment glioma MRI dataset. Challenge competitors will develop automated segmentation models to predict four distinct tumor sub-regions consisting of enhancing tissue (ET), surrounding non-enhancing T2/fluid-attenuated inversion recovery (FLAIR) hyperintensity (SNFH), non-enhancing tumor core (NETC), and resection cavity (RC). Models will be evaluated on separate validation and test datasets using standardized performance metrics utilized across the BraTS 2024 cluster of challenges, including lesion-wise Dice Similarity Coefficient and Hausdorff Distance. Models developed during this challenge will advance the field of automated MRI segmentation and contribute to their integration into clinical practice, ultimately enhancing patient care.
MedYOLO: A Medical Image Object Detection Framework
Dec 12, 2023Abstract:Artificial intelligence-enhanced identification of organs, lesions, and other structures in medical imaging is typically done using convolutional neural networks (CNNs) designed to make voxel-accurate segmentations of the region of interest. However, the labels required to train these CNNs are time-consuming to generate and require attention from subject matter experts to ensure quality. For tasks where voxel-level precision is not required, object detection models offer a viable alternative that can reduce annotation effort. Despite this potential application, there are few options for general purpose object detection frameworks available for 3-D medical imaging. We report on MedYOLO, a 3-D object detection framework using the one-shot detection method of the YOLO family of models and designed for use with medical imaging. We tested this model on four different datasets: BRaTS, LIDC, an abdominal organ Computed Tomography (CT) dataset, and an ECG-gated heart CT dataset. We found our models achieve high performance on commonly present medium and large-sized structures such as the heart, liver, and pancreas even without hyperparameter tuning. However, the models struggle with very small or rarely present structures.
The Brain Tumor Segmentation (BraTS-METS) Challenge 2023: Brain Metastasis Segmentation on Pre-treatment MRI
Jun 01, 2023



Abstract:Clinical monitoring of metastatic disease to the brain can be a laborious and time-consuming process, especially in cases involving multiple metastases when the assessment is performed manually. The Response Assessment in Neuro-Oncology Brain Metastases (RANO-BM) guideline, which utilizes the unidimensional longest diameter, is commonly used in clinical and research settings to evaluate response to therapy in patients with brain metastases. However, accurate volumetric assessment of the lesion and surrounding peri-lesional edema holds significant importance in clinical decision-making and can greatly enhance outcome prediction. The unique challenge in performing segmentations of brain metastases lies in their common occurrence as small lesions. Detection and segmentation of lesions that are smaller than 10 mm in size has not demonstrated high accuracy in prior publications. The brain metastases challenge sets itself apart from previously conducted MICCAI challenges on glioma segmentation due to the significant variability in lesion size. Unlike gliomas, which tend to be larger on presentation scans, brain metastases exhibit a wide range of sizes and tend to include small lesions. We hope that the BraTS-METS dataset and challenge will advance the field of automated brain metastasis detection and segmentation.
The Brain Tumor Segmentation Challenge 2023: Brain MR Image Synthesis for Tumor Segmentation
May 20, 2023
Abstract:Automated brain tumor segmentation methods are well established, reaching performance levels with clear clinical utility. Most algorithms require four input magnetic resonance imaging (MRI) modalities, typically T1-weighted images with and without contrast enhancement, T2-weighted images, and FLAIR images. However, some of these sequences are often missing in clinical practice, e.g., because of time constraints and/or image artifacts (such as patient motion). Therefore, substituting missing modalities to recover segmentation performance in these scenarios is highly desirable and necessary for the more widespread adoption of such algorithms in clinical routine. In this work, we report the set-up of the Brain MR Image Synthesis Benchmark (BraSyn), organized in conjunction with the Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2023. The objective of the challenge is to benchmark image synthesis methods that realistically synthesize missing MRI modalities given multiple available images to facilitate automated brain tumor segmentation pipelines. The image dataset is multi-modal and diverse, created in collaboration with various hospitals and research institutions.
SOUP-GAN: Super-Resolution MRI Using Generative Adversarial Networks
Jun 04, 2021
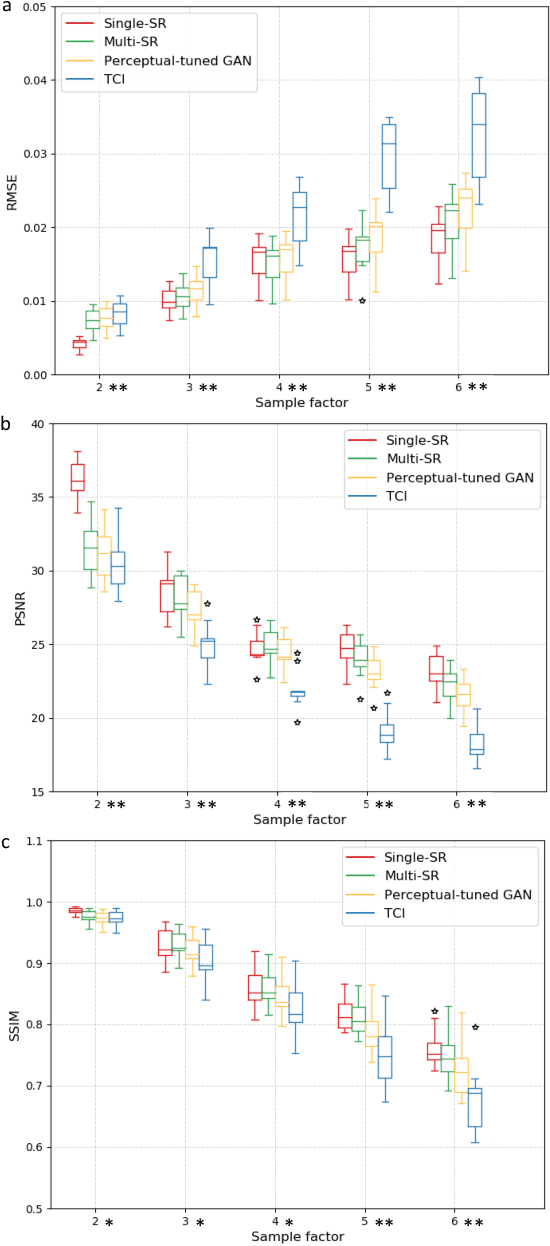
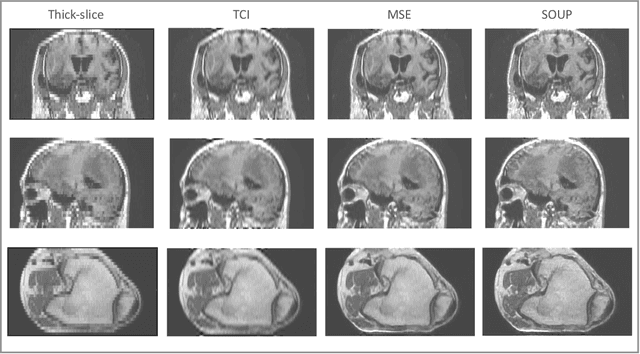
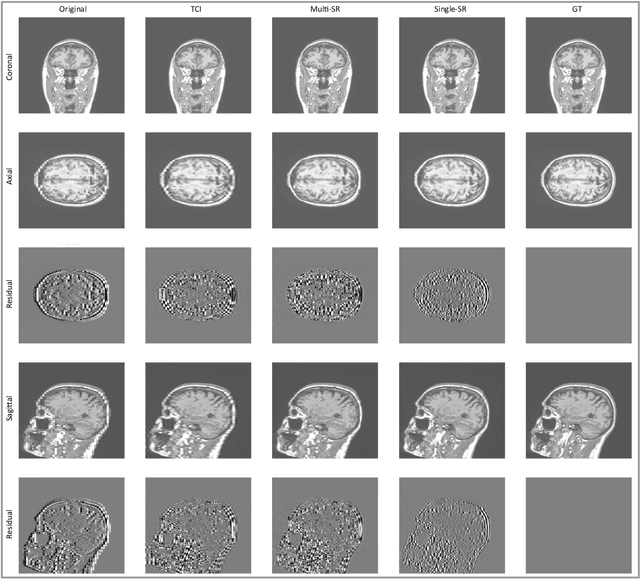
Abstract:There is a growing demand for high-resolution (HR) medical images in both the clinical and research applications. Image quality is inevitably traded off with the acquisition time for better patient comfort, lower examination costs, dose, and fewer motion-induced artifacts. For many image-based tasks, increasing the apparent resolution in the perpendicular plane to produce multi-planar reformats or 3D images is commonly used. Single image super-resolution (SR) is a promising technique to provide HR images based on unsupervised learning to increase resolution of a 2D image, but there are few reports on 3D SR. Further, perceptual loss is proposed in the literature to better capture the textual details and edges than using pixel-wise loss functions, by comparing the semantic distances in the high-dimensional feature space of a pre-trained 2D network (e.g., VGG). However, it is not clear how one should generalize it to 3D medical images, and the attendant implications are still unclear. In this paper, we propose a framework called SOUP-GAN: Super-resolution Optimized Using Perceptual-tuned Generative Adversarial Network (GAN), in order to produce thinner slice (e.g., high resolution in the 'Z' plane) medical images with anti-aliasing and deblurring. The proposed method outperforms other conventional resolution-enhancement methods and previous SR work on medical images upon both qualitative and quantitative comparisons. Specifically, we examine the model in terms of its generalization for various SR ratios and imaging modalities. By addressing those limitations, our model shows promise as a novel 3D SR interpolation technique, providing potential applications in both clinical and research settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge