Pouria Rouzrokh
LatteReview: A Multi-Agent Framework for Systematic Review Automation Using Large Language Models
Jan 05, 2025

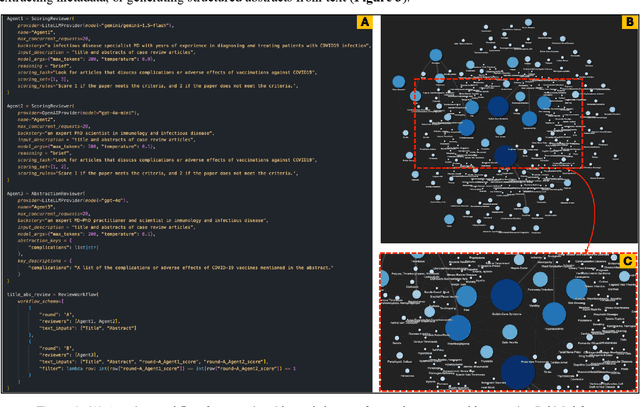
Abstract:Systematic literature reviews and meta-analyses are essential for synthesizing research insights, but they remain time-intensive and labor-intensive due to the iterative processes of screening, evaluation, and data extraction. This paper introduces and evaluates LatteReview, a Python-based framework that leverages large language models (LLMs) and multi-agent systems to automate key elements of the systematic review process. Designed to streamline workflows while maintaining rigor, LatteReview utilizes modular agents for tasks such as title and abstract screening, relevance scoring, and structured data extraction. These agents operate within orchestrated workflows, supporting sequential and parallel review rounds, dynamic decision-making, and iterative refinement based on user feedback. LatteReview's architecture integrates LLM providers, enabling compatibility with both cloud-based and locally hosted models. The framework supports features such as Retrieval-Augmented Generation (RAG) for incorporating external context, multimodal reviews, Pydantic-based validation for structured inputs and outputs, and asynchronous programming for handling large-scale datasets. The framework is available on the GitHub repository, with detailed documentation and an installable package.
RadRotator: 3D Rotation of Radiographs with Diffusion Models
Apr 19, 2024Abstract:Transforming two-dimensional (2D) images into three-dimensional (3D) volumes is a well-known yet challenging problem for the computer vision community. In the medical domain, a few previous studies attempted to convert two or more input radiographs into computed tomography (CT) volumes. Following their effort, we introduce a diffusion model-based technology that can rotate the anatomical content of any input radiograph in 3D space, potentially enabling the visualization of the entire anatomical content of the radiograph from any viewpoint in 3D. Similar to previous studies, we used CT volumes to create Digitally Reconstructed Radiographs (DRRs) as the training data for our model. However, we addressed two significant limitations encountered in previous studies: 1. We utilized conditional diffusion models with classifier-free guidance instead of Generative Adversarial Networks (GANs) to achieve higher mode coverage and improved output image quality, with the only trade-off being slower inference time, which is often less critical in medical applications; and 2. We demonstrated that the unreliable output of style transfer deep learning (DL) models, such as Cycle-GAN, to transfer the style of actual radiographs to DRRs could be replaced with a simple yet effective training transformation that randomly changes the pixel intensity histograms of the input and ground-truth imaging data during training. This transformation makes the diffusion model agnostic to any distribution variations of the input data pixel intensity, enabling the reliable training of a DL model on input DRRs and applying the exact same model to conventional radiographs (or DRRs) during inference.
CONFLARE: CONFormal LArge language model REtrieval
Apr 04, 2024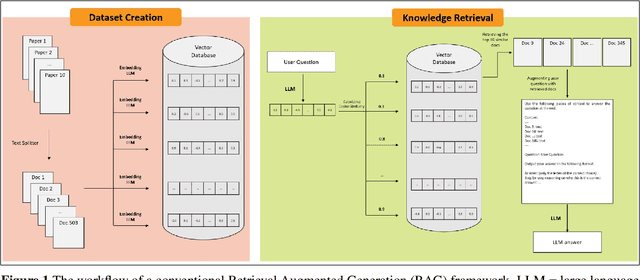
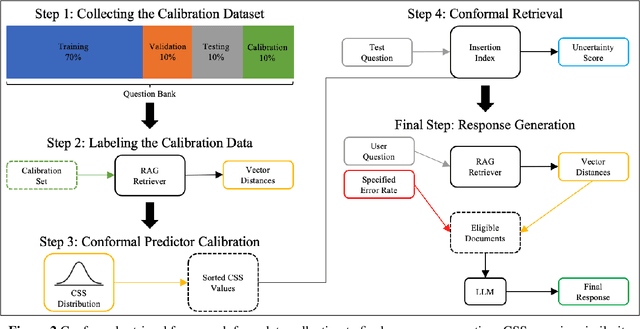
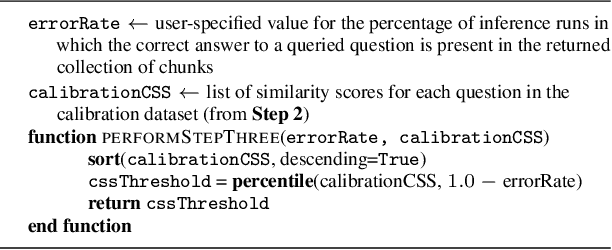

Abstract:Retrieval-augmented generation (RAG) frameworks enable large language models (LLMs) to retrieve relevant information from a knowledge base and incorporate it into the context for generating responses. This mitigates hallucinations and allows for the updating of knowledge without retraining the LLM. However, RAG does not guarantee valid responses if retrieval fails to identify the necessary information as the context for response generation. Also, if there is contradictory content, the RAG response will likely reflect only one of the two possible responses. Therefore, quantifying uncertainty in the retrieval process is crucial for ensuring RAG trustworthiness. In this report, we introduce a four-step framework for applying conformal prediction to quantify retrieval uncertainty in RAG frameworks. First, a calibration set of questions answerable from the knowledge base is constructed. Each question's embedding is compared against document embeddings to identify the most relevant document chunks containing the answer and record their similarity scores. Given a user-specified error rate ({\alpha}), these similarity scores are then analyzed to determine a similarity score cutoff threshold. During inference, all chunks with similarity exceeding this threshold are retrieved to provide context to the LLM, ensuring the true answer is captured in the context with a (1-{\alpha}) confidence level. We provide a Python package that enables users to implement the entire workflow proposed in our work, only using LLMs and without human intervention.
RIDGE: Reproducibility, Integrity, Dependability, Generalizability, and Efficiency Assessment of Medical Image Segmentation Models
Jan 16, 2024
Abstract:Deep learning techniques, despite their potential, often suffer from a lack of reproducibility and generalizability, impeding their clinical adoption. Image segmentation is one of the critical tasks in medical image analysis, in which one or several regions/volumes of interest should be annotated. This paper introduces the RIDGE checklist, a framework for assessing the Reproducibility, Integrity, Dependability, Generalizability, and Efficiency of deep learning-based medical image segmentation models. The checklist serves as a guide for researchers to enhance the quality and transparency of their work, ensuring that segmentation models are not only scientifically sound but also clinically relevant.
Synthetically Enhanced: Unveiling Synthetic Data's Potential in Medical Imaging Research
Nov 15, 2023



Abstract:Chest X-rays (CXR) are the most common medical imaging study and are used to diagnose multiple medical conditions. This study examines the impact of synthetic data supplementation, using diffusion models, on the performance of deep learning (DL) classifiers for CXR analysis. We employed three datasets: CheXpert, MIMIC-CXR, and Emory Chest X-ray, training conditional denoising diffusion probabilistic models (DDPMs) to generate synthetic frontal radiographs. Our approach ensured that synthetic images mirrored the demographic and pathological traits of the original data. Evaluating the classifiers' performance on internal and external datasets revealed that synthetic data supplementation enhances model accuracy, particularly in detecting less prevalent pathologies. Furthermore, models trained on synthetic data alone approached the performance of those trained on real data. This suggests that synthetic data can potentially compensate for real data shortages in training robust DL models. However, despite promising outcomes, the superiority of real data persists.
SOUP-GAN: Super-Resolution MRI Using Generative Adversarial Networks
Jun 04, 2021
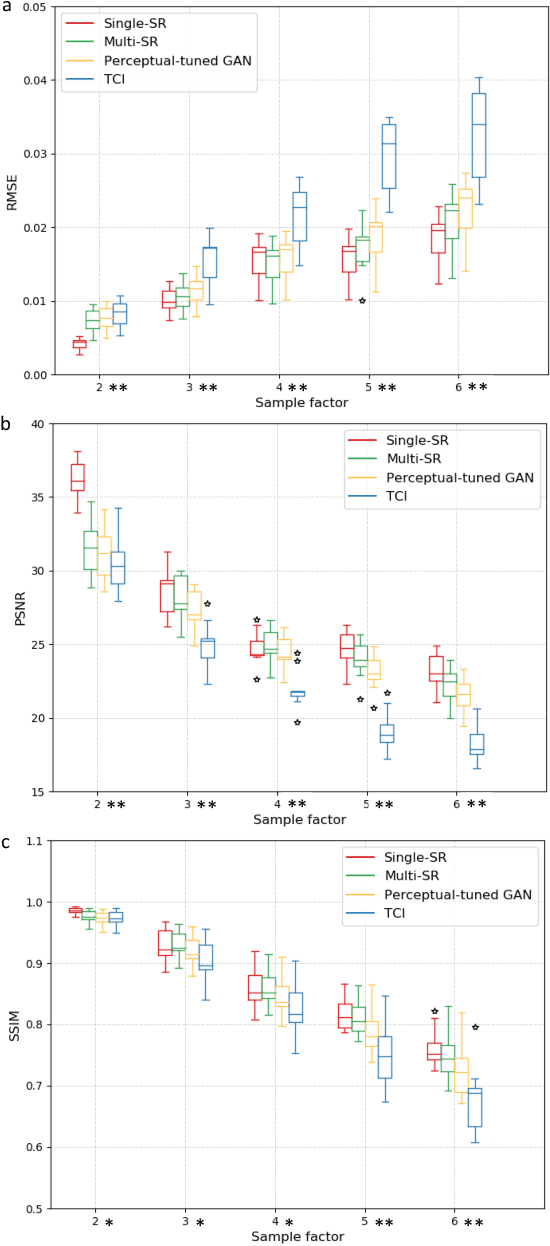
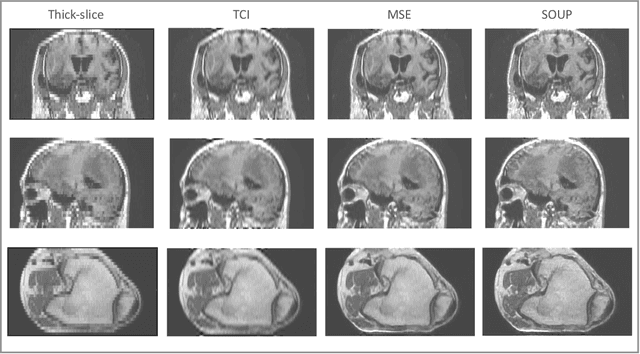
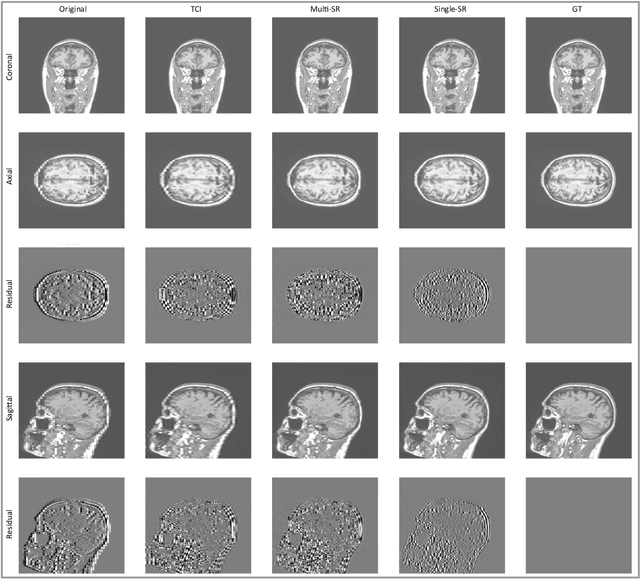
Abstract:There is a growing demand for high-resolution (HR) medical images in both the clinical and research applications. Image quality is inevitably traded off with the acquisition time for better patient comfort, lower examination costs, dose, and fewer motion-induced artifacts. For many image-based tasks, increasing the apparent resolution in the perpendicular plane to produce multi-planar reformats or 3D images is commonly used. Single image super-resolution (SR) is a promising technique to provide HR images based on unsupervised learning to increase resolution of a 2D image, but there are few reports on 3D SR. Further, perceptual loss is proposed in the literature to better capture the textual details and edges than using pixel-wise loss functions, by comparing the semantic distances in the high-dimensional feature space of a pre-trained 2D network (e.g., VGG). However, it is not clear how one should generalize it to 3D medical images, and the attendant implications are still unclear. In this paper, we propose a framework called SOUP-GAN: Super-resolution Optimized Using Perceptual-tuned Generative Adversarial Network (GAN), in order to produce thinner slice (e.g., high resolution in the 'Z' plane) medical images with anti-aliasing and deblurring. The proposed method outperforms other conventional resolution-enhancement methods and previous SR work on medical images upon both qualitative and quantitative comparisons. Specifically, we examine the model in terms of its generalization for various SR ratios and imaging modalities. By addressing those limitations, our model shows promise as a novel 3D SR interpolation technique, providing potential applications in both clinical and research settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge