Shahriar Faghani
CONRep: Uncertainty-Aware Vision-Language Report Drafting Using Conformal Prediction
Feb 03, 2026Abstract:Automated radiology report drafting (ARRD) using vision-language models (VLMs) has advanced rapidly, yet most systems lack explicit uncertainty estimates, limiting trust and safe clinical deployment. We propose CONRep, a model-agnostic framework that integrates conformal prediction (CP) to provide statistically grounded uncertainty quantification for VLM-generated radiology reports. CONRep operates at both the label level, by calibrating binary predictions for predefined findings, and the sentence level, by assessing uncertainty in free-text impressions via image-text semantic alignment. We evaluate CONRep using both generative and contrastive VLMs on public chest X-ray datasets. Across both settings, outputs classified as high confidence consistently show significantly higher agreement with radiologist annotations and ground-truth impressions than low-confidence outputs. By enabling calibrated confidence stratification without modifying underlying models, CONRep improves the transparency, reliability, and clinical usability of automated radiology reporting systems.
Analysis of the MICCAI Brain Tumor Segmentation -- Metastases (BraTS-METS) 2025 Lighthouse Challenge: Brain Metastasis Segmentation on Pre- and Post-treatment MRI
Apr 16, 2025Abstract:Despite continuous advancements in cancer treatment, brain metastatic disease remains a significant complication of primary cancer and is associated with an unfavorable prognosis. One approach for improving diagnosis, management, and outcomes is to implement algorithms based on artificial intelligence for the automated segmentation of both pre- and post-treatment MRI brain images. Such algorithms rely on volumetric criteria for lesion identification and treatment response assessment, which are still not available in clinical practice. Therefore, it is critical to establish tools for rapid volumetric segmentations methods that can be translated to clinical practice and that are trained on high quality annotated data. The BraTS-METS 2025 Lighthouse Challenge aims to address this critical need by establishing inter-rater and intra-rater variability in dataset annotation by generating high quality annotated datasets from four individual instances of segmentation by neuroradiologists while being recorded on video (two instances doing "from scratch" and two instances after AI pre-segmentation). This high-quality annotated dataset will be used for testing phase in 2025 Lighthouse challenge and will be publicly released at the completion of the challenge. The 2025 Lighthouse challenge will also release the 2023 and 2024 segmented datasets that were annotated using an established pipeline of pre-segmentation, student annotation, two neuroradiologists checking, and one neuroradiologist finalizing the process. It builds upon its previous edition by including post-treatment cases in the dataset. Using these high-quality annotated datasets, the 2025 Lighthouse challenge plans to test benchmark algorithms for automated segmentation of pre-and post-treatment brain metastases (BM), trained on diverse and multi-institutional datasets of MRI images obtained from patients with brain metastases.
CONSeg: Voxelwise Glioma Conformal Segmentation
Feb 28, 2025Abstract:Background and Purpose: Glioma segmentation is crucial for clinical decisions and treatment planning. Uncertainty quantification methods, including conformal prediction (CP), can enhance segmentation models reliability. This study aims to use CP in glioma segmentation. Methods: We used the UCSF and UPenn glioma datasets, with the UCSF dataset split into training (70%), validation (10%), calibration (10%), and test (10%) sets, and the UPenn dataset divided into external calibration (30%) and external test (70%) sets. A UNet model was trained, and its optimal threshold was set to 0.5 using prediction normalization. To apply CP, the conformal threshold was selected based on the internal/external calibration nonconformity score, and CP was subsequently applied to the internal/external test sets, with coverage reported for all. We defined the uncertainty ratio (UR) and assessed its correlation with the Dice score coefficient (DSC). Additionally, we categorized cases into certain and uncertain groups based on UR and compared their DSC. We also evaluate the correlation between UR and DSC of the BraTS fusion model segmentation (BFMS), and compare DSC in the certain and uncertain subgroups. Results: The base model achieved a DSC of 0.8628 and 0.8257 on the internal and external test sets, respectively. The CP coverage was 0.9982 for the internal test set and 0.9977 for the external test set. Statistical analysis showed a significant negative correlation between UR and DSC for test sets (p<0.001). UR was also linked to significantly lower DSCs in the BFMS (p<0.001). Additionally, certain cases had significantly higher DSCs than uncertain cases in test sets and the BFMS (p<0.001). Conclusion: CP effectively quantifies uncertainty in glioma segmentation. Using CONSeg improves the reliability of segmentation models and enhances human-computer interaction.
Comparative Analysis of 2D and 3D ResNet Architectures for IDH and MGMT Mutation Detection in Glioma Patients
Dec 30, 2024



Abstract:Gliomas are the most common cause of mortality among primary brain tumors. Molecular markers, including Isocitrate Dehydrogenase (IDH) and O[6]-methylguanine-DNA methyltransferase (MGMT) influence treatment responses and prognosis. Deep learning (DL) models may provide a non-invasive method for predicting the status of these molecular markers. To achieve non-invasive determination of gene mutations in glioma patients, we compare 2D and 3D ResNet models to predict IDH and MGMT status, using T1, post-contrast T1, and FLAIR MRI sequences. USCF glioma dataset was used, which contains 495 patients with known IDH and 410 patients with known MGMT status. The dataset was divided into training (60%), tuning (20%), and test (20%) subsets at the patient level. The 2D models take axial, coronal, and sagittal tumor slices as three separate models. To ensemble the 2D predictions the three different views were combined using logistic regression. Various ResNet architectures (ResNet10, 18, 34, 50, 101, 152) were trained. For the 3D approach, we incorporated the entire brain tumor volume in the ResNet10, 18, and 34 models. After optimizing each model, the models with the lowest tuning loss were selected for further evaluation on the separate test sets. The best-performing models in IDH prediction were the 2D ResNet50, achieving a test area under the receiver operating characteristic curve (AUROC) of 0.9096, and the 3D ResNet34, which reached a test AUROC of 0.8999. For MGMT status prediction, the 2D ResNet152 achieved a test AUROC of 0.6168; however, all 3D models yielded AUROCs less than 0.5. Overall, the study indicated that both 2D and 3D models showed high predictive value for IDH prediction, with slightly better performance in 2D models.
Brain Tumor Segmentation (BraTS) Challenge 2024: Meningioma Radiotherapy Planning Automated Segmentation
May 28, 2024Abstract:The 2024 Brain Tumor Segmentation Meningioma Radiotherapy (BraTS-MEN-RT) challenge aims to advance automated segmentation algorithms using the largest known multi-institutional dataset of radiotherapy planning brain MRIs with expert-annotated target labels for patients with intact or post-operative meningioma that underwent either conventional external beam radiotherapy or stereotactic radiosurgery. Each case includes a defaced 3D post-contrast T1-weighted radiotherapy planning MRI in its native acquisition space, accompanied by a single-label "target volume" representing the gross tumor volume (GTV) and any at-risk post-operative site. Target volume annotations adhere to established radiotherapy planning protocols, ensuring consistency across cases and institutions. For pre-operative meningiomas, the target volume encompasses the entire GTV and associated nodular dural tail, while for post-operative cases, it includes at-risk resection cavity margins as determined by the treating institution. Case annotations were reviewed and approved by expert neuroradiologists and radiation oncologists. Participating teams will develop, containerize, and evaluate automated segmentation models using this comprehensive dataset. Model performance will be assessed using the lesion-wise Dice Similarity Coefficient and the 95% Hausdorff distance. The top-performing teams will be recognized at the Medical Image Computing and Computer Assisted Intervention Conference in October 2024. BraTS-MEN-RT is expected to significantly advance automated radiotherapy planning by enabling precise tumor segmentation and facilitating tailored treatment, ultimately improving patient outcomes.
The 2024 Brain Tumor Segmentation (BraTS) Challenge: Glioma Segmentation on Post-treatment MRI
May 28, 2024
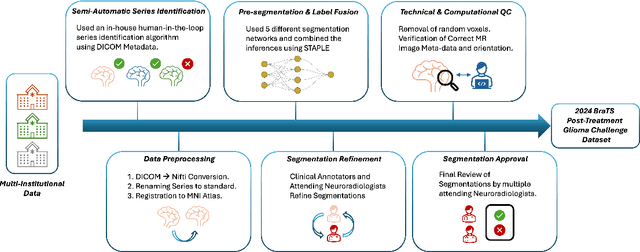
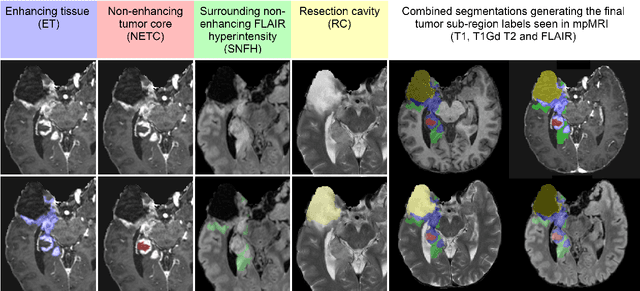
Abstract:Gliomas are the most common malignant primary brain tumors in adults and one of the deadliest types of cancer. There are many challenges in treatment and monitoring due to the genetic diversity and high intrinsic heterogeneity in appearance, shape, histology, and treatment response. Treatments include surgery, radiation, and systemic therapies, with magnetic resonance imaging (MRI) playing a key role in treatment planning and post-treatment longitudinal assessment. The 2024 Brain Tumor Segmentation (BraTS) challenge on post-treatment glioma MRI will provide a community standard and benchmark for state-of-the-art automated segmentation models based on the largest expert-annotated post-treatment glioma MRI dataset. Challenge competitors will develop automated segmentation models to predict four distinct tumor sub-regions consisting of enhancing tissue (ET), surrounding non-enhancing T2/fluid-attenuated inversion recovery (FLAIR) hyperintensity (SNFH), non-enhancing tumor core (NETC), and resection cavity (RC). Models will be evaluated on separate validation and test datasets using standardized performance metrics utilized across the BraTS 2024 cluster of challenges, including lesion-wise Dice Similarity Coefficient and Hausdorff Distance. Models developed during this challenge will advance the field of automated MRI segmentation and contribute to their integration into clinical practice, ultimately enhancing patient care.
BraTS-Path Challenge: Assessing Heterogeneous Histopathologic Brain Tumor Sub-regions
May 17, 2024Abstract:Glioblastoma is the most common primary adult brain tumor, with a grim prognosis - median survival of 12-18 months following treatment, and 4 months otherwise. Glioblastoma is widely infiltrative in the cerebral hemispheres and well-defined by heterogeneous molecular and micro-environmental histopathologic profiles, which pose a major obstacle in treatment. Correctly diagnosing these tumors and assessing their heterogeneity is crucial for choosing the precise treatment and potentially enhancing patient survival rates. In the gold-standard histopathology-based approach to tumor diagnosis, detecting various morpho-pathological features of distinct histology throughout digitized tissue sections is crucial. Such "features" include the presence of cellular tumor, geographic necrosis, pseudopalisading necrosis, areas abundant in microvascular proliferation, infiltration into the cortex, wide extension in subcortical white matter, leptomeningeal infiltration, regions dense with macrophages, and the presence of perivascular or scattered lymphocytes. With these features in mind and building upon the main aim of the BraTS Cluster of Challenges https://www.synapse.org/brats2024, the goal of the BraTS-Path challenge is to provide a systematically prepared comprehensive dataset and a benchmarking environment to develop and fairly compare deep-learning models capable of identifying tumor sub-regions of distinct histologic profile. These models aim to further our understanding of the disease and assist in the diagnosis and grading of conditions in a consistent manner.
Analysis of the BraTS 2023 Intracranial Meningioma Segmentation Challenge
May 16, 2024



Abstract:We describe the design and results from the BraTS 2023 Intracranial Meningioma Segmentation Challenge. The BraTS Meningioma Challenge differed from prior BraTS Glioma challenges in that it focused on meningiomas, which are typically benign extra-axial tumors with diverse radiologic and anatomical presentation and a propensity for multiplicity. Nine participating teams each developed deep-learning automated segmentation models using image data from the largest multi-institutional systematically expert annotated multilabel multi-sequence meningioma MRI dataset to date, which included 1000 training set cases, 141 validation set cases, and 283 hidden test set cases. Each case included T2, T2/FLAIR, T1, and T1Gd brain MRI sequences with associated tumor compartment labels delineating enhancing tumor, non-enhancing tumor, and surrounding non-enhancing T2/FLAIR hyperintensity. Participant automated segmentation models were evaluated and ranked based on a scoring system evaluating lesion-wise metrics including dice similarity coefficient (DSC) and 95% Hausdorff Distance. The top ranked team had a lesion-wise median dice similarity coefficient (DSC) of 0.976, 0.976, and 0.964 for enhancing tumor, tumor core, and whole tumor, respectively and a corresponding average DSC of 0.899, 0.904, and 0.871, respectively. These results serve as state-of-the-art benchmarks for future pre-operative meningioma automated segmentation algorithms. Additionally, we found that 1286 of 1424 cases (90.3%) had at least 1 compartment voxel abutting the edge of the skull-stripped image edge, which requires further investigation into optimal pre-processing face anonymization steps.
RadRotator: 3D Rotation of Radiographs with Diffusion Models
Apr 19, 2024Abstract:Transforming two-dimensional (2D) images into three-dimensional (3D) volumes is a well-known yet challenging problem for the computer vision community. In the medical domain, a few previous studies attempted to convert two or more input radiographs into computed tomography (CT) volumes. Following their effort, we introduce a diffusion model-based technology that can rotate the anatomical content of any input radiograph in 3D space, potentially enabling the visualization of the entire anatomical content of the radiograph from any viewpoint in 3D. Similar to previous studies, we used CT volumes to create Digitally Reconstructed Radiographs (DRRs) as the training data for our model. However, we addressed two significant limitations encountered in previous studies: 1. We utilized conditional diffusion models with classifier-free guidance instead of Generative Adversarial Networks (GANs) to achieve higher mode coverage and improved output image quality, with the only trade-off being slower inference time, which is often less critical in medical applications; and 2. We demonstrated that the unreliable output of style transfer deep learning (DL) models, such as Cycle-GAN, to transfer the style of actual radiographs to DRRs could be replaced with a simple yet effective training transformation that randomly changes the pixel intensity histograms of the input and ground-truth imaging data during training. This transformation makes the diffusion model agnostic to any distribution variations of the input data pixel intensity, enabling the reliable training of a DL model on input DRRs and applying the exact same model to conventional radiographs (or DRRs) during inference.
CONFLARE: CONFormal LArge language model REtrieval
Apr 04, 2024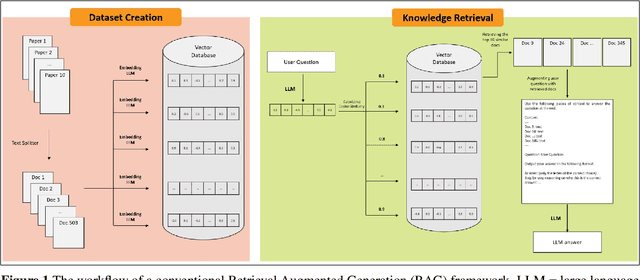
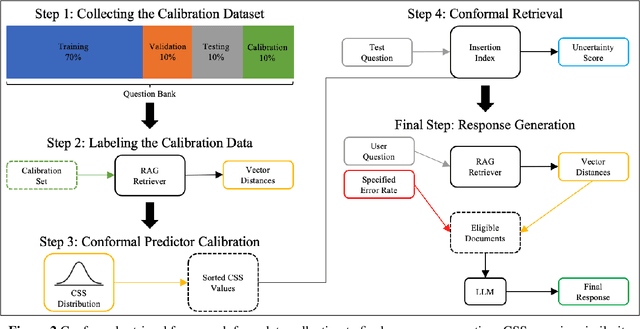
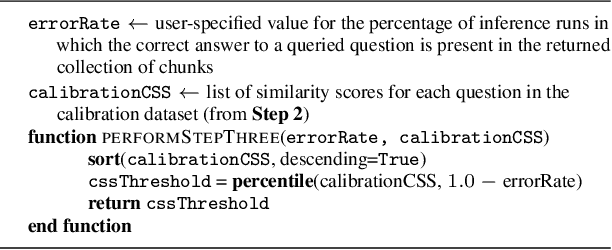

Abstract:Retrieval-augmented generation (RAG) frameworks enable large language models (LLMs) to retrieve relevant information from a knowledge base and incorporate it into the context for generating responses. This mitigates hallucinations and allows for the updating of knowledge without retraining the LLM. However, RAG does not guarantee valid responses if retrieval fails to identify the necessary information as the context for response generation. Also, if there is contradictory content, the RAG response will likely reflect only one of the two possible responses. Therefore, quantifying uncertainty in the retrieval process is crucial for ensuring RAG trustworthiness. In this report, we introduce a four-step framework for applying conformal prediction to quantify retrieval uncertainty in RAG frameworks. First, a calibration set of questions answerable from the knowledge base is constructed. Each question's embedding is compared against document embeddings to identify the most relevant document chunks containing the answer and record their similarity scores. Given a user-specified error rate ({\alpha}), these similarity scores are then analyzed to determine a similarity score cutoff threshold. During inference, all chunks with similarity exceeding this threshold are retrieved to provide context to the LLM, ensuring the true answer is captured in the context with a (1-{\alpha}) confidence level. We provide a Python package that enables users to implement the entire workflow proposed in our work, only using LLMs and without human intervention.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge