Reza Forghani
WaveFormer: A 3D Transformer with Wavelet-Driven Feature Representation for Efficient Medical Image Segmentation
Apr 01, 2025Abstract:Transformer-based architectures have advanced medical image analysis by effectively modeling long-range dependencies, yet they often struggle in 3D settings due to substantial memory overhead and insufficient capture of fine-grained local features. We address these limitations with WaveFormer, a novel 3D-transformer that: i) leverages the fundamental frequency-domain properties of features for contextual representation, and ii) is inspired by the top-down mechanism of the human visual recognition system, making it a biologically motivated architecture. By employing discrete wavelet transformations (DWT) at multiple scales, WaveFormer preserves both global context and high-frequency details while replacing heavy upsampling layers with efficient wavelet-based summarization and reconstruction. This significantly reduces the number of parameters, which is critical for real-world deployment where computational resources and training times are constrained. Furthermore, the model is generic and easily adaptable to diverse applications. Evaluations on BraTS2023, FLARE2021, and KiTS2023 demonstrate performance on par with state-of-the-art methods while offering substantially lower computational complexity.
RIDGE: Reproducibility, Integrity, Dependability, Generalizability, and Efficiency Assessment of Medical Image Segmentation Models
Jan 16, 2024
Abstract:Deep learning techniques, despite their potential, often suffer from a lack of reproducibility and generalizability, impeding their clinical adoption. Image segmentation is one of the critical tasks in medical image analysis, in which one or several regions/volumes of interest should be annotated. This paper introduces the RIDGE checklist, a framework for assessing the Reproducibility, Integrity, Dependability, Generalizability, and Efficiency of deep learning-based medical image segmentation models. The checklist serves as a guide for researchers to enhance the quality and transparency of their work, ensuring that segmentation models are not only scientifically sound but also clinically relevant.
Generalizability of Machine Learning Models: Quantitative Evaluation of Three Methodological Pitfalls
Feb 01, 2022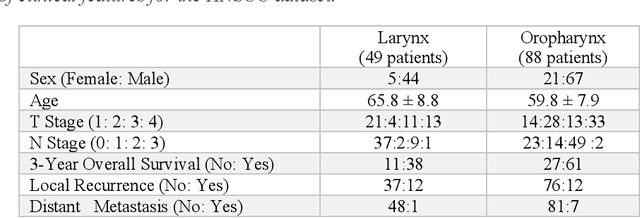
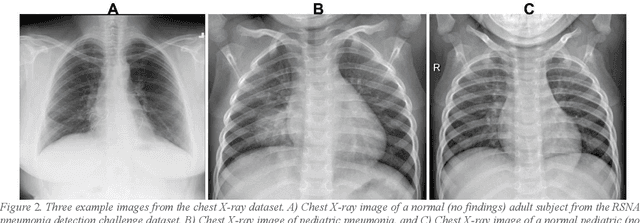
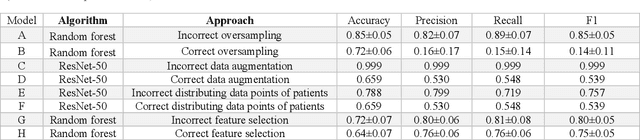
Abstract:Despite the great potential of machine learning, the lack of generalizability has hindered the widespread adoption of these technologies in routine clinical practice. We investigate three methodological pitfalls: (1) violation of independence assumption, (2) model evaluation with an inappropriate performance indicator, and (3) batch effect and how these pitfalls could affect the generalizability of machine learning models. We implement random forest and deep convolutional neural network models using several medical imaging datasets, including head and neck CT, lung CT, chest X-Ray, and histopathological images, to quantify and illustrate the effect of these pitfalls. We develop these models with and without the pitfall and compare the performance of the resulting models in terms of accuracy, precision, recall, and F1 score. Our results showed that violation of the independence assumption could substantially affect model generalizability. More specifically, (I) applying oversampling before splitting data into train, validation and test sets; (II) performing data augmentation before splitting data; (III) distributing data points for a subject across training, validation, and test sets; and (IV) applying feature selection before splitting data led to superficial boosts in model performance. We also observed that inappropriate performance indicators could lead to erroneous conclusions. Also, batch effect could lead to developing models that lack generalizability. The aforementioned methodological pitfalls lead to machine learning models with over-optimistic performance. These errors, if made, cannot be captured using internal model evaluation, and the inaccurate predictions made by the model may lead to wrong conclusions and interpretations. Therefore, avoiding these pitfalls is a necessary condition for developing generalizable models.
Spectral image clustering on dual-energy CT scans using functional regression mixtures
Jan 31, 2022



Abstract:Dual-energy computed tomography (DECT) is an advanced CT scanning technique enabling material characterization not possible with conventional CT scans. It allows the reconstruction of energy decay curves at each 3D image voxel, representing varying image attenuation at different effective scanning energy levels. In this paper, we develop novel functional data analysis (FDA) techniques and adapt them to the analysis of DECT decay curves. More specifically, we construct functional mixture models that integrate spatial context in mixture weights, with mixture component densities being constructed upon the energy decay curves as functional observations. We design unsupervised clustering algorithms by developing dedicated expectation maximization (EM) algorithms for the maximum likelihood estimation of the model parameters. To our knowledge, this is the first article to adapt statistical FDA tools and model-based clustering to take advantage of the full spectral information provided by DECT. We evaluate our methods on 91 head and neck cancer DECT scans. We compare our unsupervised clustering results to tumor contours traced manually by radiologists, as well as to several baseline algorithms. Given the inter-rater variability even among experts at delineating head and neck tumors, and given the potential importance of tissue reactions surrounding the tumor itself, our proposed methodology has the potential to add value in downstream machine learning applications for clinical outcome prediction based on DECT data in head and neck cancer.
Does Proprietary Software Still Offer Protection of Intellectual Property in the Age of Machine Learning? -- A Case Study using Dual Energy CT Data
Dec 06, 2021



Abstract:In the domain of medical image processing, medical device manufacturers protect their intellectual property in many cases by shipping only compiled software, i.e. binary code which can be executed but is difficult to be understood by a potential attacker. In this paper, we investigate how well this procedure is able to protect image processing algorithms. In particular, we investigate whether the computation of mono-energetic images and iodine maps from dual energy CT data can be reverse-engineered by machine learning methods. Our results indicate that both can be approximated using only one single slice image as training data at a very high accuracy with structural similarity greater than 0.98 in all investigated cases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge