Rajiv Gupta
Generalizability of Machine Learning Models: Quantitative Evaluation of Three Methodological Pitfalls
Feb 01, 2022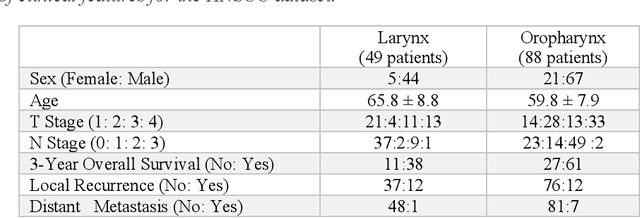
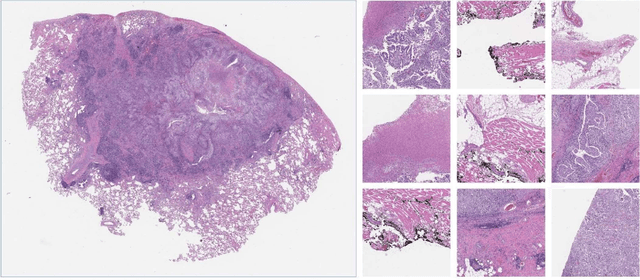
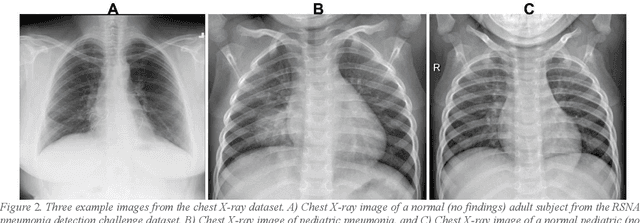
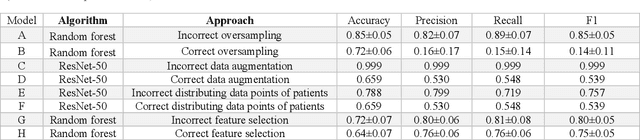
Abstract:Despite the great potential of machine learning, the lack of generalizability has hindered the widespread adoption of these technologies in routine clinical practice. We investigate three methodological pitfalls: (1) violation of independence assumption, (2) model evaluation with an inappropriate performance indicator, and (3) batch effect and how these pitfalls could affect the generalizability of machine learning models. We implement random forest and deep convolutional neural network models using several medical imaging datasets, including head and neck CT, lung CT, chest X-Ray, and histopathological images, to quantify and illustrate the effect of these pitfalls. We develop these models with and without the pitfall and compare the performance of the resulting models in terms of accuracy, precision, recall, and F1 score. Our results showed that violation of the independence assumption could substantially affect model generalizability. More specifically, (I) applying oversampling before splitting data into train, validation and test sets; (II) performing data augmentation before splitting data; (III) distributing data points for a subject across training, validation, and test sets; and (IV) applying feature selection before splitting data led to superficial boosts in model performance. We also observed that inappropriate performance indicators could lead to erroneous conclusions. Also, batch effect could lead to developing models that lack generalizability. The aforementioned methodological pitfalls lead to machine learning models with over-optimistic performance. These errors, if made, cannot be captured using internal model evaluation, and the inaccurate predictions made by the model may lead to wrong conclusions and interpretations. Therefore, avoiding these pitfalls is a necessary condition for developing generalizable models.
Urine Microscopic Image Dataset
Nov 19, 2021Abstract:Urinalysis is a standard diagnostic test to detect urinary system related problems. The automation of urinalysis will reduce the overall diagnostic time. Recent studies used urine microscopic datasets for designing deep learning based algorithms to classify and detect urine cells. But these datasets are not publicly available for further research. To alleviate the need for urine datsets, we prepare our urine sediment microscopic image (UMID) dataset comprising of around 3700 cell annotations and 3 categories of cells namely RBC, pus and epithelial cells. We discuss the several challenges involved in preparing the dataset and the annotations. We make the dataset publicly available.
Split Learning for collaborative deep learning in healthcare
Dec 27, 2019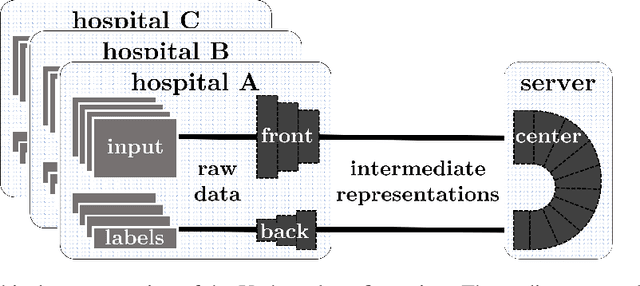
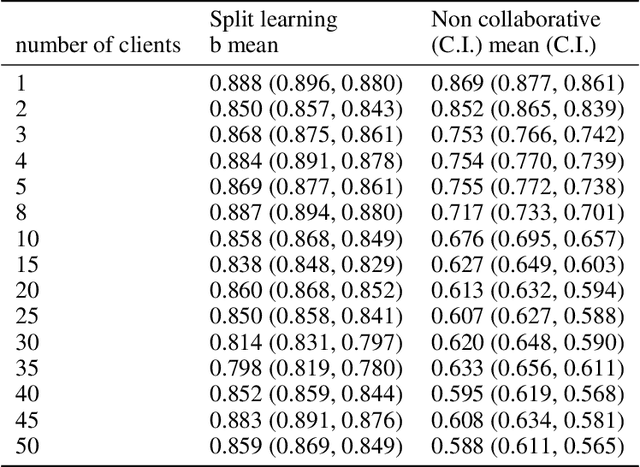
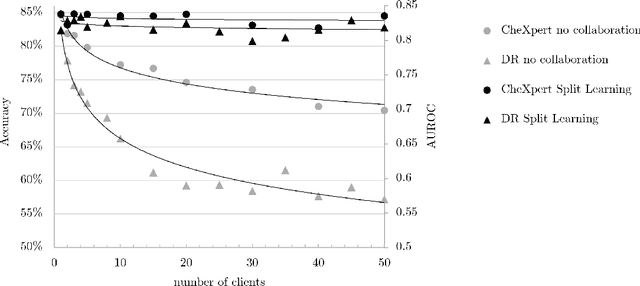
Abstract:Shortage of labeled data has been holding the surge of deep learning in healthcare back, as sample sizes are often small, patient information cannot be shared openly, and multi-center collaborative studies are a burden to set up. Distributed machine learning methods promise to mitigate these problems. We argue for a split learning based approach and apply this distributed learning method for the first time in the medical field to compare performance against (1) centrally hosted and (2) non collaborative configurations for a range of participants. Two medical deep learning tasks are used to compare split learning to conventional single and multi center approaches: a binary classification problem of a data set of 9000 fundus photos, and multi-label classification problem of a data set of 156,535 chest X-rays. The several distributed learning setups are compared for a range of 1-50 distributed participants. Performance of the split learning configuration remained constant for any number of clients compared to a single center study, showing a marked difference compared to the non collaborative configuration after 2 clients (p < 0.001) for both sets. Our results affirm the benefits of collaborative training of deep neural networks in health care. Our work proves the significant benefit of distributed learning in healthcare, and paves the way for future real-world implementations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge