Katie Ovens
Rel-UNet: Reliable Tumor Segmentation via Uncertainty Quantification in nnU-Net
Mar 11, 2025



Abstract:Accurate and reliable tumor segmentation is essential in medical imaging analysis for improving diagnosis, treatment planning, and monitoring. However, existing segmentation models often lack robust mechanisms for quantifying the uncertainty associated with their predictions, which is essential for informed clinical decision-making. This study presents a novel approach for uncertainty quantification in kidney tumor segmentation using deep learning, specifically leveraging multiple local minima during training. Our method generates uncertainty maps without modifying the original model architecture or requiring extensive computational resources. We evaluated our approach on the KiTS23 dataset, where our approach effectively identified ambiguous regions faster and with lower uncertainty scores in contrast to previous approaches. The generated uncertainty maps provide critical insights into model confidence, ultimately enhancing the reliability of the segmentation with the potential to support more accurate medical diagnoses. The computational efficiency and model-agnostic design of the proposed approach allows adaptation without architectural changes, enabling use across various segmentation models.
Modified CycleGAN for the synthesization of samples for wheat head segmentation
Feb 23, 2024



Abstract:Deep learning models have been used for a variety of image processing tasks. However, most of these models are developed through supervised learning approaches, which rely heavily on the availability of large-scale annotated datasets. Developing such datasets is tedious and expensive. In the absence of an annotated dataset, synthetic data can be used for model development; however, due to the substantial differences between simulated and real data, a phenomenon referred to as domain gap, the resulting models often underperform when applied to real data. In this research, we aim to address this challenge by first computationally simulating a large-scale annotated dataset and then using a generative adversarial network (GAN) to fill the gap between simulated and real images. This approach results in a synthetic dataset that can be effectively utilized to train a deep-learning model. Using this approach, we developed a realistic annotated synthetic dataset for wheat head segmentation. This dataset was then used to develop a deep-learning model for semantic segmentation. The resulting model achieved a Dice score of 83.4\% on an internal dataset and Dice scores of 79.6% and 83.6% on two external Global Wheat Head Detection datasets. While we proposed this approach in the context of wheat head segmentation, it can be generalized to other crop types or, more broadly, to images with dense, repeated patterns such as those found in cellular imagery.
RIDGE: Reproducibility, Integrity, Dependability, Generalizability, and Efficiency Assessment of Medical Image Segmentation Models
Jan 16, 2024
Abstract:Deep learning techniques, despite their potential, often suffer from a lack of reproducibility and generalizability, impeding their clinical adoption. Image segmentation is one of the critical tasks in medical image analysis, in which one or several regions/volumes of interest should be annotated. This paper introduces the RIDGE checklist, a framework for assessing the Reproducibility, Integrity, Dependability, Generalizability, and Efficiency of deep learning-based medical image segmentation models. The checklist serves as a guide for researchers to enhance the quality and transparency of their work, ensuring that segmentation models are not only scientifically sound but also clinically relevant.
Generalizability of Machine Learning Models: Quantitative Evaluation of Three Methodological Pitfalls
Feb 01, 2022
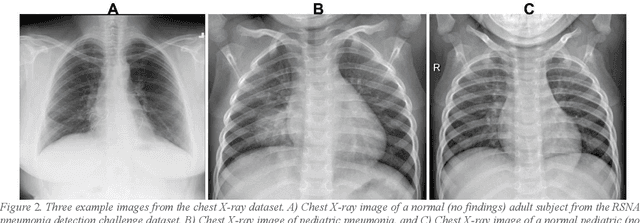
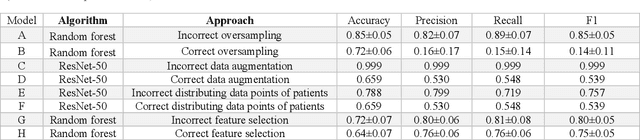
Abstract:Despite the great potential of machine learning, the lack of generalizability has hindered the widespread adoption of these technologies in routine clinical practice. We investigate three methodological pitfalls: (1) violation of independence assumption, (2) model evaluation with an inappropriate performance indicator, and (3) batch effect and how these pitfalls could affect the generalizability of machine learning models. We implement random forest and deep convolutional neural network models using several medical imaging datasets, including head and neck CT, lung CT, chest X-Ray, and histopathological images, to quantify and illustrate the effect of these pitfalls. We develop these models with and without the pitfall and compare the performance of the resulting models in terms of accuracy, precision, recall, and F1 score. Our results showed that violation of the independence assumption could substantially affect model generalizability. More specifically, (I) applying oversampling before splitting data into train, validation and test sets; (II) performing data augmentation before splitting data; (III) distributing data points for a subject across training, validation, and test sets; and (IV) applying feature selection before splitting data led to superficial boosts in model performance. We also observed that inappropriate performance indicators could lead to erroneous conclusions. Also, batch effect could lead to developing models that lack generalizability. The aforementioned methodological pitfalls lead to machine learning models with over-optimistic performance. These errors, if made, cannot be captured using internal model evaluation, and the inaccurate predictions made by the model may lead to wrong conclusions and interpretations. Therefore, avoiding these pitfalls is a necessary condition for developing generalizable models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge