Steve Langer
RIDGE: Reproducibility, Integrity, Dependability, Generalizability, and Efficiency Assessment of Medical Image Segmentation Models
Jan 16, 2024
Abstract:Deep learning techniques, despite their potential, often suffer from a lack of reproducibility and generalizability, impeding their clinical adoption. Image segmentation is one of the critical tasks in medical image analysis, in which one or several regions/volumes of interest should be annotated. This paper introduces the RIDGE checklist, a framework for assessing the Reproducibility, Integrity, Dependability, Generalizability, and Efficiency of deep learning-based medical image segmentation models. The checklist serves as a guide for researchers to enhance the quality and transparency of their work, ensuring that segmentation models are not only scientifically sound but also clinically relevant.
Best Practices and Scoring System on Reviewing A.I. based Medical Imaging Papers: Part 1 Classification
Feb 03, 2022
Abstract:With the recent advances in A.I. methodologies and their application to medical imaging, there has been an explosion of related research programs utilizing these techniques to produce state-of-the-art classification performance. Ultimately, these research programs culminate in submission of their work for consideration in peer reviewed journals. To date, the criteria for acceptance vs. rejection is often subjective; however, reproducible science requires reproducible review. The Machine Learning Education Sub-Committee of SIIM has identified a knowledge gap and a serious need to establish guidelines for reviewing these studies. Although there have been several recent papers with this goal, this present work is written from the machine learning practitioners standpoint. In this series, the committee will address the best practices to be followed in an A.I.-based study and present the required sections in terms of examples and discussion of what should be included to make the studies cohesive, reproducible, accurate, and self-contained. This first entry in the series focuses on the task of image classification. Elements such as dataset curation, data pre-processing steps, defining an appropriate reference standard, data partitioning, model architecture and training are discussed. The sections are presented as they would be detailed in a typical manuscript, with content describing the necessary information that should be included to make sure the study is of sufficient quality to be considered for publication. The goal of this series is to provide resources to not only help improve the review process for A.I.-based medical imaging papers, but to facilitate a standard for the information that is presented within all components of the research study. We hope to provide quantitative metrics in what otherwise may be a qualitative review process.
A Patient-Centric Dataset of Images and Metadata for Identifying Melanomas Using Clinical Context
Aug 07, 2020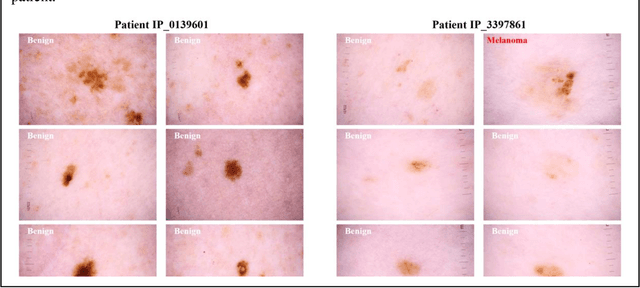
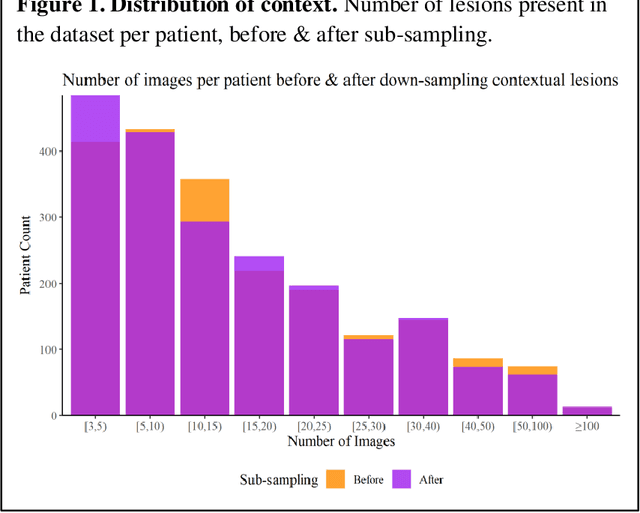
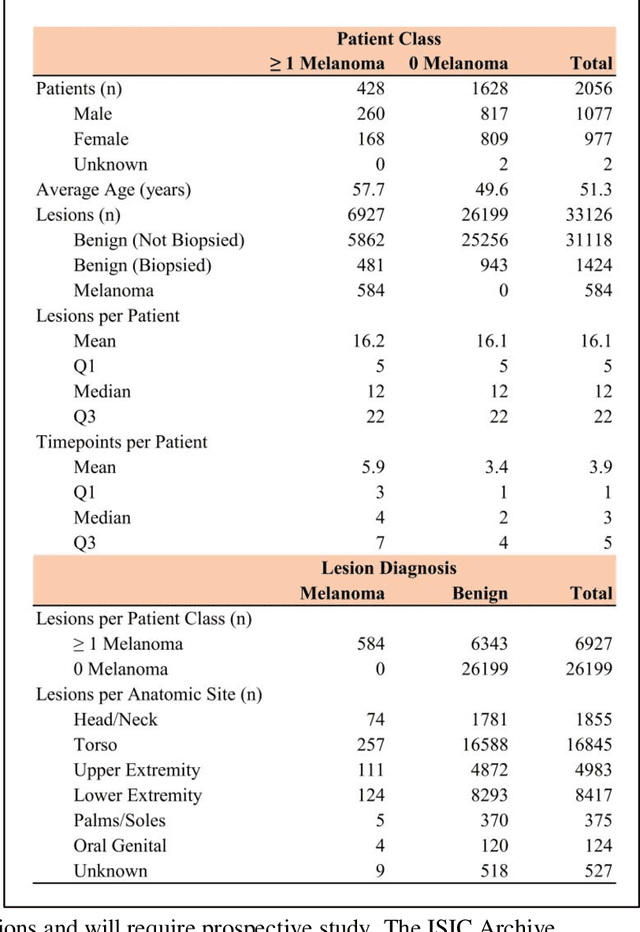
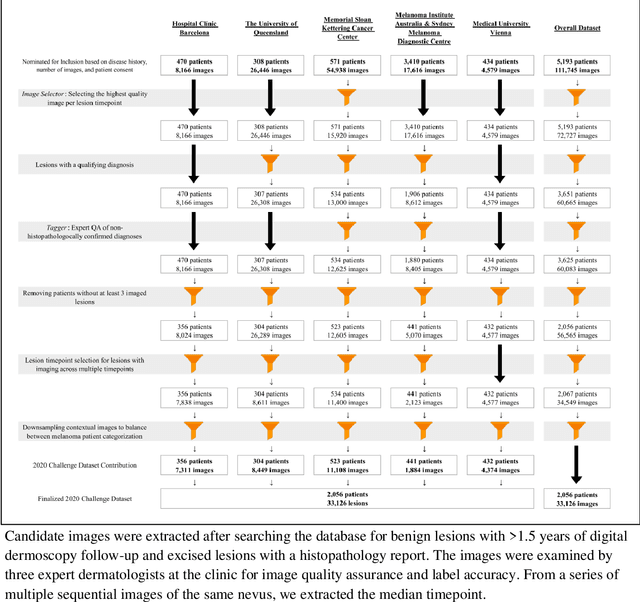
Abstract:Prior skin image datasets have not addressed patient-level information obtained from multiple skin lesions from the same patient. Though artificial intelligence classification algorithms have achieved expert-level performance in controlled studies examining single images, in practice dermatologists base their judgment holistically from multiple lesions on the same patient. The 2020 SIIM-ISIC Melanoma Classification challenge dataset described herein was constructed to address this discrepancy between prior challenges and clinical practice, providing for each image in the dataset an identifier allowing lesions from the same patient to be mapped to one another. This patient-level contextual information is frequently used by clinicians to diagnose melanoma and is especially useful in ruling out false positives in patients with many atypical nevi. The dataset represents 2,056 patients from three continents with an average of 16 lesions per patient, consisting of 33,126 dermoscopic images and 584 histopathologically confirmed melanomas compared with benign melanoma mimickers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge