Felipe Kitamura
The RSNA Lumbar Degenerative Imaging Spine Classification (LumbarDISC) Dataset
Jun 10, 2025Abstract:The Radiological Society of North America (RSNA) Lumbar Degenerative Imaging Spine Classification (LumbarDISC) dataset is the largest publicly available dataset of adult MRI lumbar spine examinations annotated for degenerative changes. The dataset includes 2,697 patients with a total of 8,593 image series from 8 institutions across 6 countries and 5 continents. The dataset is available for free for non-commercial use via Kaggle and RSNA Medical Imaging Resource of AI (MIRA). The dataset was created for the RSNA 2024 Lumbar Spine Degenerative Classification competition where competitors developed deep learning models to grade degenerative changes in the lumbar spine. The degree of spinal canal, subarticular recess, and neural foraminal stenosis was graded at each intervertebral disc level in the lumbar spine. The images were annotated by expert volunteer neuroradiologists and musculoskeletal radiologists from the RSNA, American Society of Neuroradiology, and the American Society of Spine Radiology. This dataset aims to facilitate research and development in machine learning and lumbar spine imaging to lead to improved patient care and clinical efficiency.
RIDGE: Reproducibility, Integrity, Dependability, Generalizability, and Efficiency Assessment of Medical Image Segmentation Models
Jan 16, 2024
Abstract:Deep learning techniques, despite their potential, often suffer from a lack of reproducibility and generalizability, impeding their clinical adoption. Image segmentation is one of the critical tasks in medical image analysis, in which one or several regions/volumes of interest should be annotated. This paper introduces the RIDGE checklist, a framework for assessing the Reproducibility, Integrity, Dependability, Generalizability, and Efficiency of deep learning-based medical image segmentation models. The checklist serves as a guide for researchers to enhance the quality and transparency of their work, ensuring that segmentation models are not only scientifically sound but also clinically relevant.
Federated Learning on Heterogenous Data using Chest CT
Mar 23, 2023Abstract:Large data have accelerated advances in AI. While it is well known that population differences from genetics, sex, race, diet, and various environmental factors contribute significantly to disease, AI studies in medicine have largely focused on locoregional patient cohorts with less diverse data sources. Such limitation stems from barriers to large-scale data share in medicine and ethical concerns over data privacy. Federated learning (FL) is one potential pathway for AI development that enables learning across hospitals without data share. In this study, we show the results of various FL strategies on one of the largest and most diverse COVID-19 chest CT datasets: 21 participating hospitals across five continents that comprise >10,000 patients with >1 million images. We present three techniques: Fed Averaging (FedAvg), Incremental Institutional Learning (IIL), and Cyclical Incremental Institutional Learning (CIIL). We also propose an FL strategy that leverages synthetically generated data to overcome class imbalances and data size disparities across centers. We show that FL can achieve comparable performance to Centralized Data Sharing (CDS) while maintaining high performance across sites with small, underrepresented data. We investigate the strengths and weaknesses for all technical approaches on this heterogeneous dataset including the robustness to non-Independent and identically distributed (non-IID) diversity of data. We also describe the sources of data heterogeneity such as age, sex, and site locations in the context of FL and show how even among the correctly labeled populations, disparities can arise due to these biases.
Best Practices and Scoring System on Reviewing A.I. based Medical Imaging Papers: Part 1 Classification
Feb 03, 2022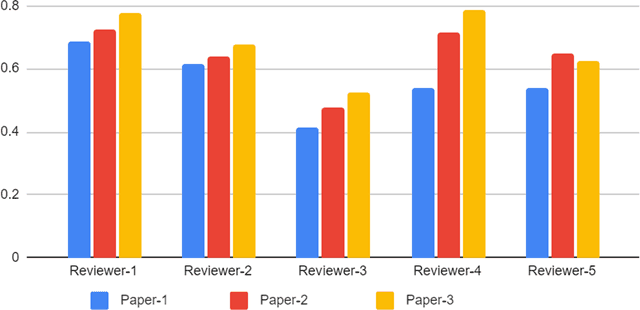
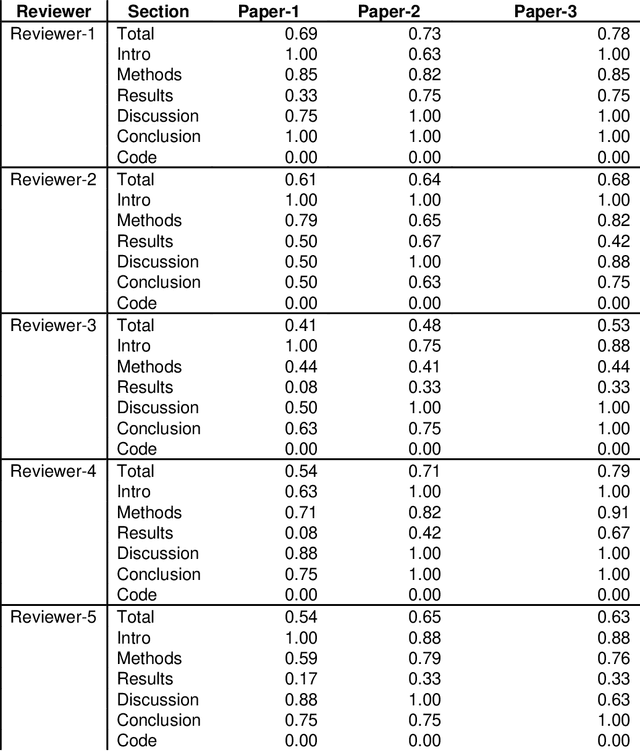
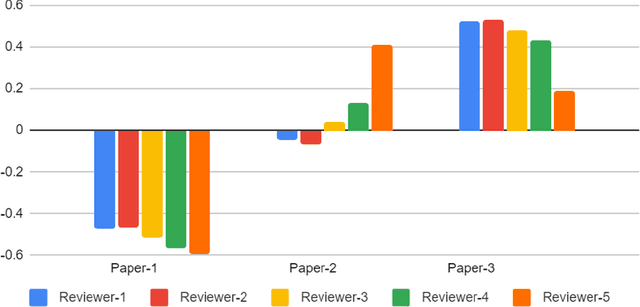
Abstract:With the recent advances in A.I. methodologies and their application to medical imaging, there has been an explosion of related research programs utilizing these techniques to produce state-of-the-art classification performance. Ultimately, these research programs culminate in submission of their work for consideration in peer reviewed journals. To date, the criteria for acceptance vs. rejection is often subjective; however, reproducible science requires reproducible review. The Machine Learning Education Sub-Committee of SIIM has identified a knowledge gap and a serious need to establish guidelines for reviewing these studies. Although there have been several recent papers with this goal, this present work is written from the machine learning practitioners standpoint. In this series, the committee will address the best practices to be followed in an A.I.-based study and present the required sections in terms of examples and discussion of what should be included to make the studies cohesive, reproducible, accurate, and self-contained. This first entry in the series focuses on the task of image classification. Elements such as dataset curation, data pre-processing steps, defining an appropriate reference standard, data partitioning, model architecture and training are discussed. The sections are presented as they would be detailed in a typical manuscript, with content describing the necessary information that should be included to make sure the study is of sufficient quality to be considered for publication. The goal of this series is to provide resources to not only help improve the review process for A.I.-based medical imaging papers, but to facilitate a standard for the information that is presented within all components of the research study. We hope to provide quantitative metrics in what otherwise may be a qualitative review process.
Federated Learning for Breast Density Classification: A Real-World Implementation
Sep 17, 2020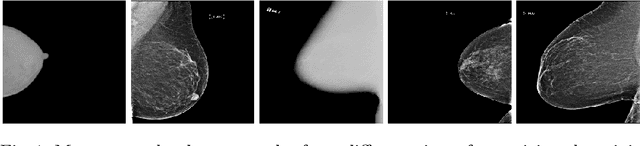

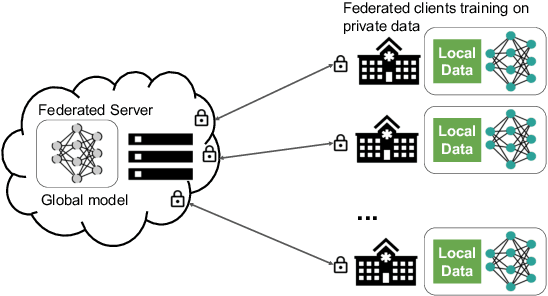
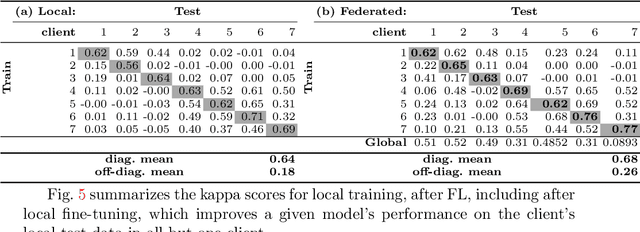
Abstract:Building robust deep learning-based models requires large quantities of diverse training data. In this study, we investigate the use of federated learning (FL) to build medical imaging classification models in a real-world collaborative setting. Seven clinical institutions from across the world joined this FL effort to train a model for breast density classification based on Breast Imaging, Reporting & Data System (BI-RADS). We show that despite substantial differences among the datasets from all sites (mammography system, class distribution, and data set size) and without centralizing data, we can successfully train AI models in federation. The results show that models trained using FL perform 6.3% on average better than their counterparts trained on an institute's local data alone. Furthermore, we show a 45.8% relative improvement in the models' generalizability when evaluated on the other participating sites' testing data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge