Ujjwal Ratan
Federated Learning on Heterogenous Data using Chest CT
Mar 23, 2023Abstract:Large data have accelerated advances in AI. While it is well known that population differences from genetics, sex, race, diet, and various environmental factors contribute significantly to disease, AI studies in medicine have largely focused on locoregional patient cohorts with less diverse data sources. Such limitation stems from barriers to large-scale data share in medicine and ethical concerns over data privacy. Federated learning (FL) is one potential pathway for AI development that enables learning across hospitals without data share. In this study, we show the results of various FL strategies on one of the largest and most diverse COVID-19 chest CT datasets: 21 participating hospitals across five continents that comprise >10,000 patients with >1 million images. We present three techniques: Fed Averaging (FedAvg), Incremental Institutional Learning (IIL), and Cyclical Incremental Institutional Learning (CIIL). We also propose an FL strategy that leverages synthetically generated data to overcome class imbalances and data size disparities across centers. We show that FL can achieve comparable performance to Centralized Data Sharing (CDS) while maintaining high performance across sites with small, underrepresented data. We investigate the strengths and weaknesses for all technical approaches on this heterogeneous dataset including the robustness to non-Independent and identically distributed (non-IID) diversity of data. We also describe the sources of data heterogeneity such as age, sex, and site locations in the context of FL and show how even among the correctly labeled populations, disparities can arise due to these biases.
The Ability of Image-Language Explainable Models to Resemble Domain Expertise
Sep 19, 2022



Abstract:Recent advances in vision and language (V+L) models have a promising impact in the healthcare field. However, such models struggle to explain how and why a particular decision was made. In addition, model transparency and involvement of domain expertise are critical success factors for machine learning models to make an entrance into the field. In this work, we study the use of the local surrogate explainability technique to overcome the problem of black-box deep learning models. We explore the feasibility of resembling domain expertise using the local surrogates in combination with an underlying V+L to generate multi-modal visual and language explanations. We demonstrate that such explanations can serve as helpful feedback in guiding model training for data scientists and machine learning engineers in the field.
Improving Early Sepsis Prediction with Multi Modal Learning
Jul 23, 2021
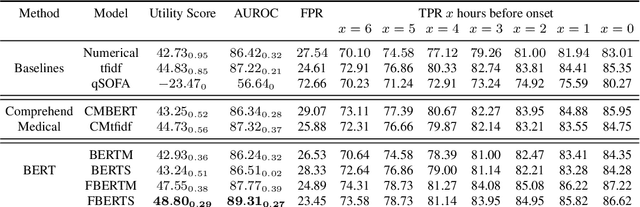


Abstract:Sepsis is a life-threatening disease with high morbidity, mortality and healthcare costs. The early prediction and administration of antibiotics and intravenous fluids is considered crucial for the treatment of sepsis and can save potentially millions of lives and billions in health care costs. Professional clinical care practitioners have proposed clinical criterion which aid in early detection of sepsis; however, performance of these criterion is often limited. Clinical text provides essential information to estimate the severity of the sepsis in addition to structured clinical data. In this study, we explore how clinical text can complement structured data towards early sepsis prediction task. In this paper, we propose multi modal model which incorporates both structured data in the form of patient measurements as well as textual notes on the patient. We employ state-of-the-art NLP models such as BERT and a highly specialized NLP model in Amazon Comprehend Medical to represent the text. On the MIMIC-III dataset containing records of ICU admissions, we show that by using these notes, one achieves an improvement of 6.07 points in a standard utility score for Sepsis prediction and 2.89% in AUROC score. Our methods significantly outperforms a clinical criteria suggested by experts, qSOFA, as well as the winning model of the PhysioNet Computing in Cardiology Challenge for predicting Sepsis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge