Fred Qin
Improving Early Sepsis Prediction with Multi Modal Learning
Jul 23, 2021
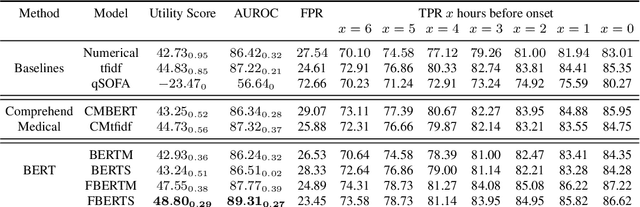


Abstract:Sepsis is a life-threatening disease with high morbidity, mortality and healthcare costs. The early prediction and administration of antibiotics and intravenous fluids is considered crucial for the treatment of sepsis and can save potentially millions of lives and billions in health care costs. Professional clinical care practitioners have proposed clinical criterion which aid in early detection of sepsis; however, performance of these criterion is often limited. Clinical text provides essential information to estimate the severity of the sepsis in addition to structured clinical data. In this study, we explore how clinical text can complement structured data towards early sepsis prediction task. In this paper, we propose multi modal model which incorporates both structured data in the form of patient measurements as well as textual notes on the patient. We employ state-of-the-art NLP models such as BERT and a highly specialized NLP model in Amazon Comprehend Medical to represent the text. On the MIMIC-III dataset containing records of ICU admissions, we show that by using these notes, one achieves an improvement of 6.07 points in a standard utility score for Sepsis prediction and 2.89% in AUROC score. Our methods significantly outperforms a clinical criteria suggested by experts, qSOFA, as well as the winning model of the PhysioNet Computing in Cardiology Challenge for predicting Sepsis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge