Sharut Gupta
ReasonCACHE: Teaching LLMs To Reason Without Weight Updates
Feb 02, 2026Abstract:Can Large language models (LLMs) learn to reason without any weight update and only through in-context learning (ICL)? ICL is strikingly sample-efficient, often learning from only a handful of demonstrations, but complex reasoning tasks typically demand many training examples to learn from. However, naively scaling ICL by adding more demonstrations breaks down at this scale: attention costs grow quadratically, performance saturates or degrades with longer contexts, and the approach remains a shallow form of learning. Due to these limitations, practitioners predominantly rely on in-weight learning (IWL) to induce reasoning. In this work, we show that by using Prefix Tuning, LLMs can learn to reason without overloading the context window and without any weight updates. We introduce $\textbf{ReasonCACHE}$, an instantiation of this mechanism that distills demonstrations into a fixed key-value cache. Empirically, across challenging reasoning benchmarks, including GPQA-Diamond, ReasonCACHE outperforms standard ICL and matches or surpasses IWL approaches. Further, it achieves this all while being more efficient across three key axes: data, inference cost, and trainable parameters. We also theoretically prove that ReasonCACHE can be strictly more expressive than low-rank weight update since the latter ties expressivity to input rank, whereas ReasonCACHE bypasses this constraint by directly injecting key-values into the attention mechanism. Together, our findings identify ReasonCACHE as a middle path between in-context and in-weight learning, providing a scalable algorithm for learning reasoning skills beyond the context window without modifying parameters. Our project page: https://reasoncache.github.io/
Sequential-Parallel Duality in Prefix Scannable Models
Jun 12, 2025Abstract:Modern neural sequence models are designed to meet the dual mandate of parallelizable training and fast sequential inference. Recent developments have given rise to various models, such as Gated Linear Attention (GLA) and Mamba, that achieve such ``sequential-parallel duality.'' This raises a natural question: can we characterize the full class of neural sequence models that support near-constant-time parallel evaluation and linear-time, constant-space sequential inference? We begin by describing a broad class of such models -- state space models -- as those whose state updates can be computed using the classic parallel prefix scan algorithm with a custom associative aggregation operator. We then define a more general class, Prefix-Scannable Models (PSMs), by relaxing the state aggregation operator to allow arbitrary (potentially non-associative) functions such as softmax attention. This generalization unifies many existing architectures, including element-wise RNNs (e.g., Mamba) and linear transformers (e.g., GLA, Mamba2, mLSTM), while also introducing new models with softmax-like operators that achieve O(1) amortized compute per token and log(N) memory for sequence length N. We empirically evaluate such models on illustrative small-scale language modeling and canonical synthetic tasks, including state tracking and associative recall. Empirically, we find that PSMs retain the expressivity of transformer-based architectures while matching the inference efficiency of state space models -- in some cases exhibiting better length generalization than either.
Understanding the Role of Equivariance in Self-supervised Learning
Nov 10, 2024Abstract:Contrastive learning has been a leading paradigm for self-supervised learning, but it is widely observed that it comes at the price of sacrificing useful features (\eg colors) by being invariant to data augmentations. Given this limitation, there has been a surge of interest in equivariant self-supervised learning (E-SSL) that learns features to be augmentation-aware. However, even for the simplest rotation prediction method, there is a lack of rigorous understanding of why, when, and how E-SSL learns useful features for downstream tasks. To bridge this gap between practice and theory, we establish an information-theoretic perspective to understand the generalization ability of E-SSL. In particular, we identify a critical explaining-away effect in E-SSL that creates a synergy between the equivariant and classification tasks. This synergy effect encourages models to extract class-relevant features to improve its equivariant prediction, which, in turn, benefits downstream tasks requiring semantic features. Based on this perspective, we theoretically analyze the influence of data transformations and reveal several principles for practical designs of E-SSL. Our theory not only aligns well with existing E-SSL methods but also sheds light on new directions by exploring the benefits of model equivariance. We believe that a theoretically grounded understanding on the role of equivariance would inspire more principled and advanced designs in this field. Code is available at https://github.com/kaotty/Understanding-ESSL.
An Information Criterion for Controlled Disentanglement of Multimodal Data
Oct 31, 2024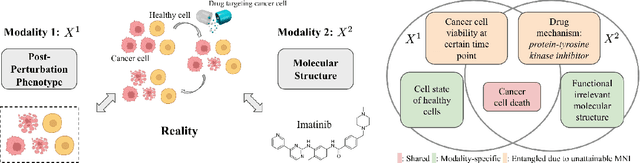
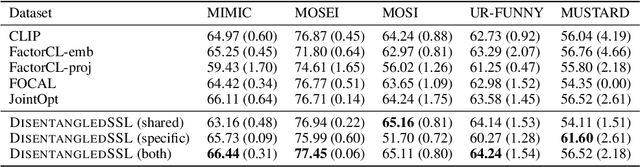
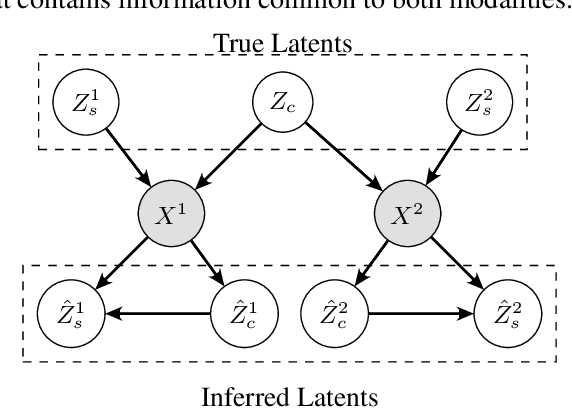
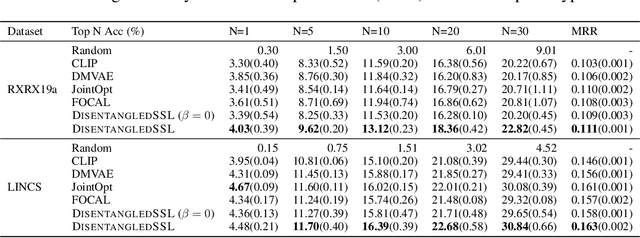
Abstract:Multimodal representation learning seeks to relate and decompose information inherent in multiple modalities. By disentangling modality-specific information from information that is shared across modalities, we can improve interpretability and robustness and enable downstream tasks such as the generation of counterfactual outcomes. Separating the two types of information is challenging since they are often deeply entangled in many real-world applications. We propose Disentangled Self-Supervised Learning (DisentangledSSL), a novel self-supervised approach for learning disentangled representations. We present a comprehensive analysis of the optimality of each disentangled representation, particularly focusing on the scenario not covered in prior work where the so-called Minimum Necessary Information (MNI) point is not attainable. We demonstrate that DisentangledSSL successfully learns shared and modality-specific features on multiple synthetic and real-world datasets and consistently outperforms baselines on various downstream tasks, including prediction tasks for vision-language data, as well as molecule-phenotype retrieval tasks for biological data.
In-Context Symmetries: Self-Supervised Learning through Contextual World Models
May 28, 2024Abstract:At the core of self-supervised learning for vision is the idea of learning invariant or equivariant representations with respect to a set of data transformations. This approach, however, introduces strong inductive biases, which can render the representations fragile in downstream tasks that do not conform to these symmetries. In this work, drawing insights from world models, we propose to instead learn a general representation that can adapt to be invariant or equivariant to different transformations by paying attention to context -- a memory module that tracks task-specific states, actions, and future states. Here, the action is the transformation, while the current and future states respectively represent the input's representation before and after the transformation. Our proposed algorithm, Contextual Self-Supervised Learning (ContextSSL), learns equivariance to all transformations (as opposed to invariance). In this way, the model can learn to encode all relevant features as general representations while having the versatility to tail down to task-wise symmetries when given a few examples as the context. Empirically, we demonstrate significant performance gains over existing methods on equivariance-related tasks, supported by both qualitative and quantitative evaluations.
Removing Biases from Molecular Representations via Information Maximization
Dec 01, 2023Abstract:High-throughput drug screening -- using cell imaging or gene expression measurements as readouts of drug effect -- is a critical tool in biotechnology to assess and understand the relationship between the chemical structure and biological activity of a drug. Since large-scale screens have to be divided into multiple experiments, a key difficulty is dealing with batch effects, which can introduce systematic errors and non-biological associations in the data. We propose InfoCORE, an Information maximization approach for COnfounder REmoval, to effectively deal with batch effects and obtain refined molecular representations. InfoCORE establishes a variational lower bound on the conditional mutual information of the latent representations given a batch identifier. It adaptively reweighs samples to equalize their implied batch distribution. Extensive experiments on drug screening data reveal InfoCORE's superior performance in a multitude of tasks including molecular property prediction and molecule-phenotype retrieval. Additionally, we show results for how InfoCORE offers a versatile framework and resolves general distribution shifts and issues of data fairness by minimizing correlation with spurious features or removing sensitive attributes. The code is available at https://github.com/uhlerlab/InfoCORE.
Context is Environment
Sep 20, 2023



Abstract:Two lines of work are taking the central stage in AI research. On the one hand, the community is making increasing efforts to build models that discard spurious correlations and generalize better in novel test environments. Unfortunately, the bitter lesson so far is that no proposal convincingly outperforms a simple empirical risk minimization baseline. On the other hand, large language models (LLMs) have erupted as algorithms able to learn in-context, generalizing on-the-fly to eclectic contextual circumstances that users enforce by means of prompting. In this paper, we argue that context is environment, and posit that in-context learning holds the key to better domain generalization. Via extensive theory and experiments, we show that paying attention to context$\unicode{x2013}\unicode{x2013}$unlabeled examples as they arrive$\unicode{x2013}\unicode{x2013}$allows our proposed In-Context Risk Minimization (ICRM) algorithm to zoom-in on the test environment risk minimizer, leading to significant out-of-distribution performance improvements. From all of this, two messages are worth taking home. Researchers in domain generalization should consider environment as context, and harness the adaptive power of in-context learning. Researchers in LLMs should consider context as environment, to better structure data towards generalization.
Structuring Representation Geometry with Rotationally Equivariant Contrastive Learning
Jun 24, 2023



Abstract:Self-supervised learning converts raw perceptual data such as images to a compact space where simple Euclidean distances measure meaningful variations in data. In this paper, we extend this formulation by adding additional geometric structure to the embedding space by enforcing transformations of input space to correspond to simple (i.e., linear) transformations of embedding space. Specifically, in the contrastive learning setting, we introduce an equivariance objective and theoretically prove that its minima forces augmentations on input space to correspond to rotations on the spherical embedding space. We show that merely combining our equivariant loss with a non-collapse term results in non-trivial representations, without requiring invariance to data augmentations. Optimal performance is achieved by also encouraging approximate invariance, where input augmentations correspond to small rotations. Our method, CARE: Contrastive Augmentation-induced Rotational Equivariance, leads to improved performance on downstream tasks, and ensures sensitivity in embedding space to important variations in data (e.g., color) that standard contrastive methods do not achieve. Code is available at https://github.com/Sharut/CARE.
FL Games: A Federated Learning Framework for Distribution Shifts
Oct 31, 2022



Abstract:Federated learning aims to train predictive models for data that is distributed across clients, under the orchestration of a server. However, participating clients typically each hold data from a different distribution, which can yield to catastrophic generalization on data from a different client, which represents a new domain. In this work, we argue that in order to generalize better across non-i.i.d. clients, it is imperative to only learn correlations that are stable and invariant across domains. We propose FL GAMES, a game-theoretic framework for federated learning that learns causal features that are invariant across clients. While training to achieve the Nash equilibrium, the traditional best response strategy suffers from high-frequency oscillations. We demonstrate that FL GAMES effectively resolves this challenge and exhibits smooth performance curves. Further, FL GAMES scales well in the number of clients, requires significantly fewer communication rounds, and is agnostic to device heterogeneity. Through empirical evaluation, we demonstrate that FL GAMES achieves high out-of-distribution performance on various benchmarks.
Minimizing Client Drift in Federated Learning via Adaptive Bias Estimation
Apr 27, 2022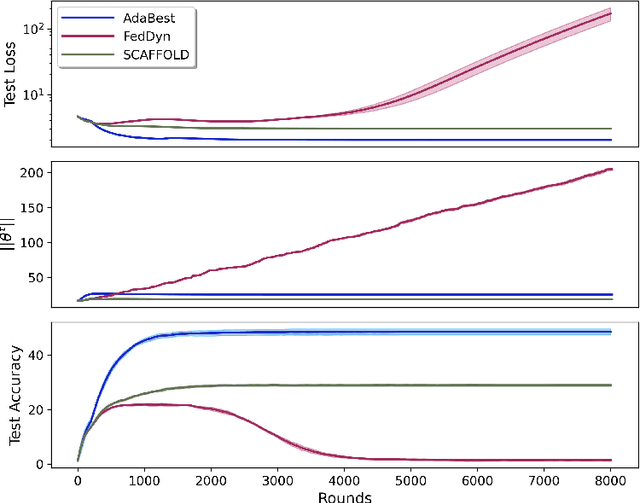

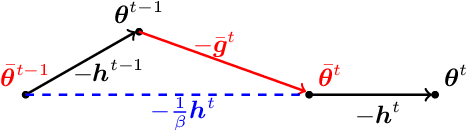
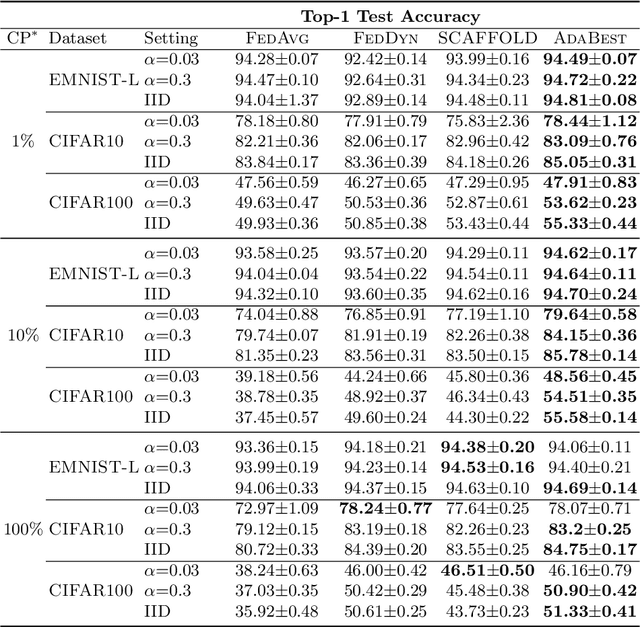
Abstract:In Federated Learning a number of clients collaborate to train a model without sharing their data. Client models are optimized locally and are communicated through a central hub called server. A major challenge is to deal with heterogeneity among clients' data which causes the local optimization to drift away with respect to the global objective. In order to estimate and therefore remove this drift, variance reduction techniques have been incorporated into Federated Learning optimization recently. However, the existing solutions propagate the error of their estimations, throughout the optimization trajectory which leads to inaccurate approximations of the clients' drift and ultimately failure to remove them properly. In this paper, we address this issue by introducing an adaptive algorithm that efficiently reduces clients' drift. Compared to the previous works on adapting variance reduction to Federated Learning, our approach uses less or the same level of communication bandwidth, computation or memory. Additionally, it addresses the instability problem--prevalent in prior work, caused by increasing norm of the estimates which makes our approach a much more practical solution for large scale Federated Learning settings. Our experimental results demonstrate that the proposed algorithm converges significantly faster and achieves higher accuracy compared to the baselines in an extensive set of Federated Learning benchmarks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge